Search Count: 40
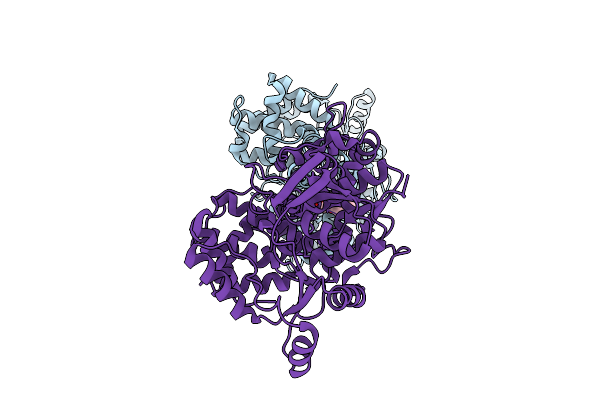 |
Crystal Structure Of Glutamate--Trna Ligase (Gltx) From Moraxella Catarrhalis (Apo)
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-12-24 Classification: LIGASE Ligands: SO4, P6G, CL |
 |
Crystal Structure Of Glutamate--Trna Ligase (Gltx) From Moraxella Catarrhalis In Complex With 5'-O-(N-Glutamyl)Sulfamoyladeonosine
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2025-10-22 Classification: LIGASE Ligands: GSU, SO4, P6G, 2PE |
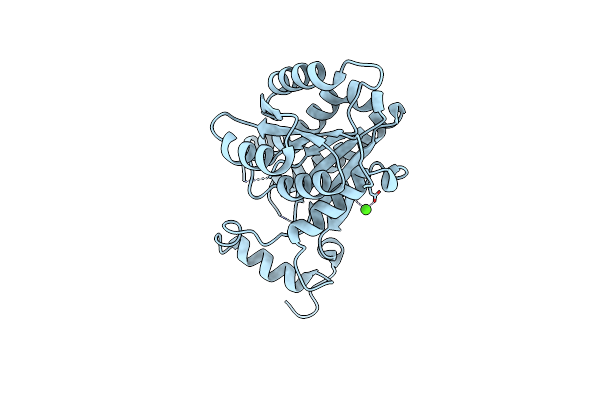 |
Crystal Structure Of The Apo Enoyl-Acp-Reductase (Fabi) From Moraxella Catarrhalis
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: CA |
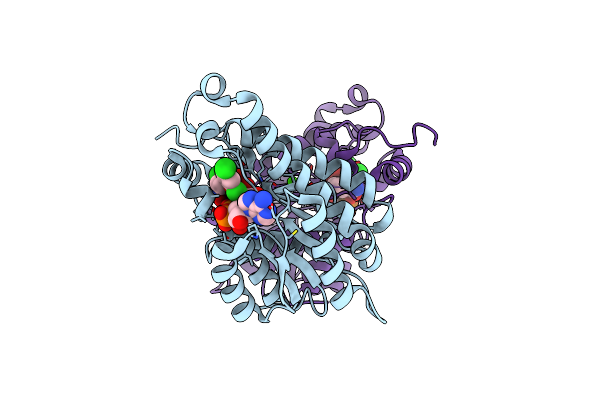 |
Crystal Structure Of Moraxella Catarrhalis Enoyl-Acp-Reductase (Fabi) In Complex With Nad And Triclosan
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-07-20 Classification: OXIDOREDUCTASE Ligands: NAD, TCL, CA |
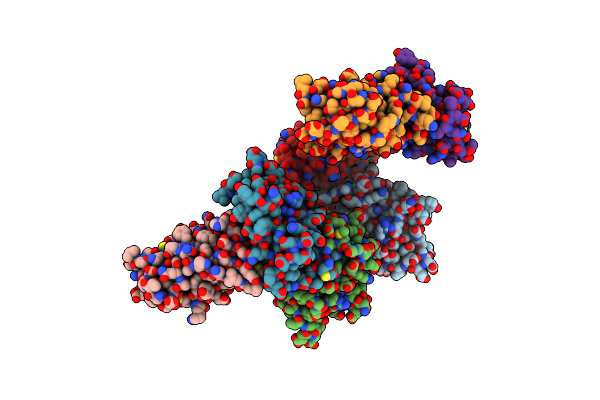 |
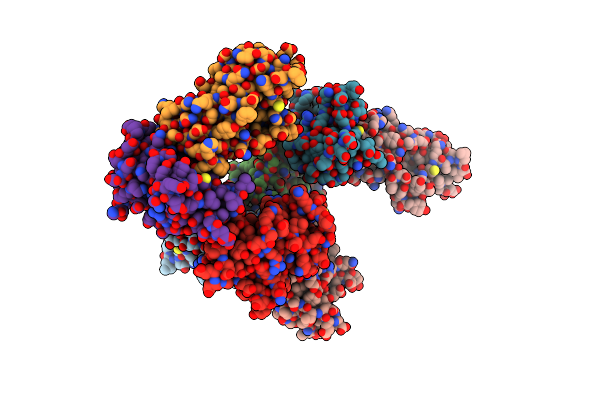
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2022-07-06 Classification: VIRAL PROTEIN |
 |
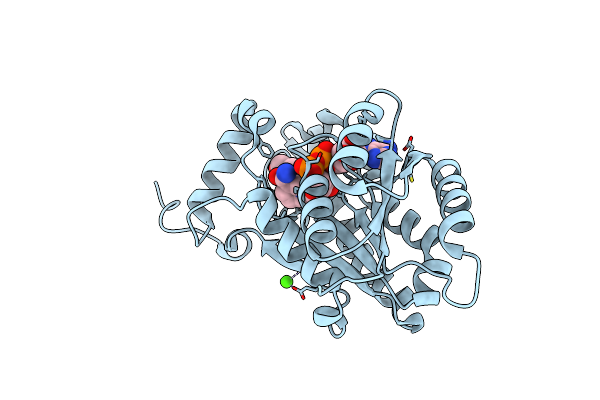
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-07-06 Classification: VIRAL PROTEIN |
 |
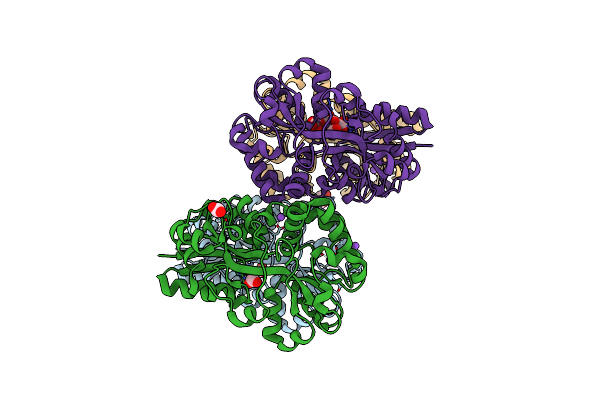
Crystal Structure Of Moraxella Catarrhalis Enoyl-Acp-Reductase (Fabi) In Complex With The Cofactor Nad
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-06-22 Classification: OXIDOREDUCTASE Ligands: NAD, CA, GOL |
 |
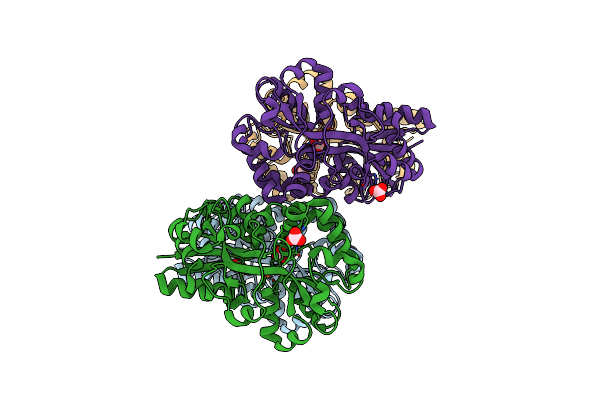
Crystal Structure Of Apo Moraxella Catarrhalis Ferric Binding Protein A In An Open Conformation
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-02-23 Classification: METAL TRANSPORT Ligands: CO3, NA, GOL, PEG, CIT |
 |
Crystal Structure Of Holo Moraxella Catarrhalis Ferric Binding Protein A In An Open Conformation
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-02-23 Classification: METAL TRANSPORT Ligands: CO3, FE |
 |
Structure Of 299-452 Fragment Of The Uspa1 Protein From Moraxella Catarrhalis
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-08-21 Classification: CELL ADHESION Ligands: CL, HEZ, ZN |
 |
Crystal Structure Of Udp-N-Acetylglucosamine O-Acyltransferase (Lpxa) From Moraxella Catarrhalis Rh4.
Organism: Moraxella catarrhalis (strain bbh18)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-06-14 Classification: TRANSFERASE Ligands: GOL, FLC |
 |
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2016-06-29 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-06-29 Classification: OXIDOREDUCTASE Ligands: MG, FOM |
 |
Crystal Structure Of The Moraxella Catarrhalis Dox-P Reductoisomerase In Complex With Nadh, Fosmidomycin And Magnesium
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-06-29 Classification: OXIDOREDUCTASE Ligands: NAD, MG, FOM, GOL, NHE |
 |
Crystal Structure Of Dox-P Reductoisomerase In Complex With Nadph, Fosmidomycin And Magnesium
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-06-29 Classification: OXIDOREDUCTASE Ligands: NDP, MG, FOM, GOL, NHE |
 |
Crystal Structure Of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase From Moraxella Catarrhalis In Complex With Magnesium Ion And Phosphate Ion
Organism: Moraxella catarrhalis bc8
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2015-02-11 Classification: HYDROLASE Ligands: MG, PO4, EDO |
 |
Crystal Structure Of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase (Kdsc) From Moraxella Catarrhalis In Complex With Magnesium Ion
Organism: Moraxella catarrhalis bc8
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2015-02-11 Classification: HYDROLASE Ligands: MG, EDO |
 |
Crystal Structure Of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase From Moraxella Catarrhalis In Complex With Citrate
Organism: Moraxella catarrhalis bc8
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-02-11 Classification: HYDROLASE Ligands: CIT |
 |
Crystal Structure Of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase From Moraxella Catarrhalis In Complex With Magnesium Ion And Kdo Molecule
Organism: Moraxella catarrhalis bc8
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2015-02-11 Classification: HYDROLASE Ligands: KDO, MG |
 |
Crystal Structure Of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase From Moraxella Catarrhalis In Complex With Magnesium Ion, Phosphate Ion And Kdo Molecule
Organism: Moraxella catarrhalis bc8
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2015-02-11 Classification: HYDROLASE Ligands: PO4, MG, KDO |

