Search Count: 188
 |
Asymmetric Dimerization In A Transcription Factor Superfamily Is Promoted By Allosteric Interactions With Dna
Organism: Mus musculus, Dna molecule
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-08-09 Classification: HORMONE Ligands: ZN |
 |
The Crystallographic Structure Of The Ligand Binding Domain Of The Nr7 Nuclear Receptor From The Amphioxus Branchiostoma Lanceolatum
Organism: Branchiostoma lanceolatum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: TRANSCRIPTION Ligands: CL, PO4 |
 |
Crystal Structure Of The Ligand-Binding Domains Of The Heterodimer Ecr/Usp Bound To The Synthetic Agonist Byi08346
Organism: Heliothis virescens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-04-07 Classification: TRANSCRIPTION Ligands: EPH, PEG, 834, PE8, PGE, MG |
 |
Crystal Structure Of The Ligand-Binding Domains Of The Heterodimer Ecr/Usp Bound To The Synthetic Agonist Byi09181
Organism: Heliothis virescens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2021-04-07 Classification: TRANSCRIPTION Ligands: EPH, U0H, MG, PEG |
 |
A Revisited Version Of The Apo Structure Of The Ligand-Binding Domain Of The Human Nuclear Receptor Rxr-Alpha
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2019-02-20 Classification: TRANSCRIPTION Ligands: CPS |
 |
Glycation Restrains Allosteric Transition In Hemoglobin: The Molecular Basis Of Oxidative Stress Under Hyperglycemic Conditions In Diabetes
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2017-02-01 Classification: OXYGEN TRANSPORT Ligands: HEM, OXY, FRU, GLC |
 |
Crystal Structure Of The Alpha Subunit Of Glycyl Trna Synthetase (Glyrs) From Aquifex Aeolicus In Complex With An Analog Of Glycyl Adenylate (Gly-Sa)
Organism: Aquifex aeolicus (strain vf5)
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-05-11 Classification: LIGASE Ligands: G5A |
 |
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-10-07 Classification: Transcription Factor/Transcription Regulator Ligands: VDX |
 |
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2015-10-07 Classification: Transcription Factor/Transcription Regulator Ligands: BIV |
 |
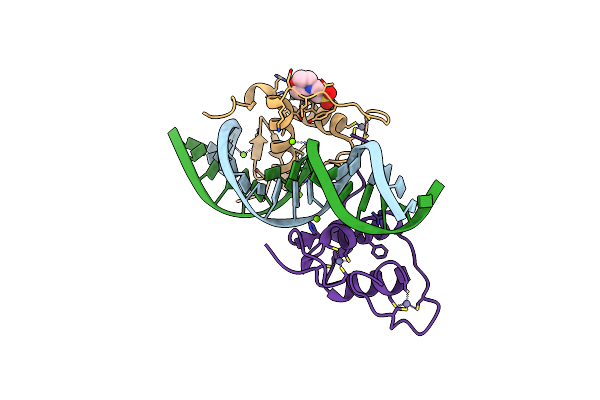
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-10-07 Classification: Transcription Factor Ligands: H97 |
 |
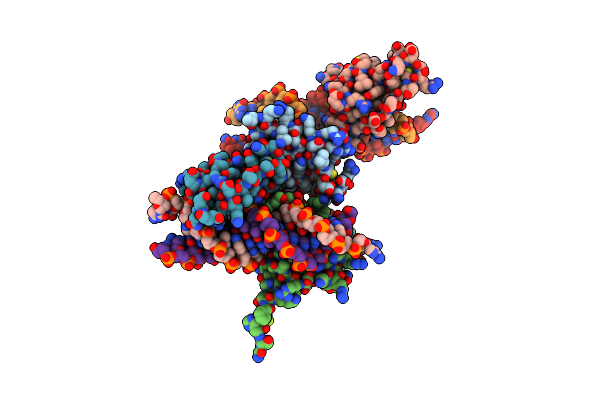
Crystal Structure Of The Human Retinoid X Receptor Dna-Binding Domain Bound To The Human Ramp2 Response Element
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2015-02-18 Classification: TRANSCRIPTION/DNA Ligands: MG, ZN, MES, CL |
 |
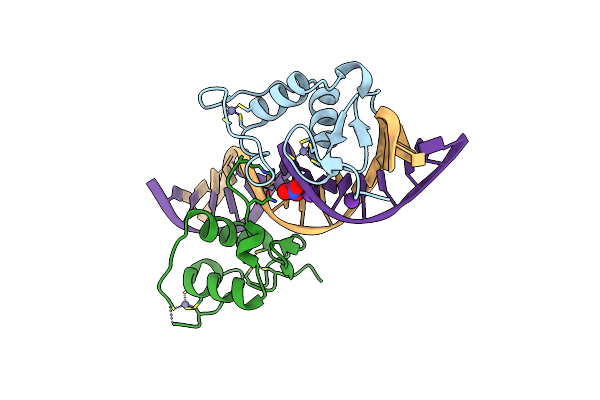
Crystal Structure Of The Human Retinoid X Receptor Dna-Binding Domain Bound To The Human Gde1Spa Response Element
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-02-18 Classification: TRANSCRIPTION/DNA Ligands: ZN |
 |
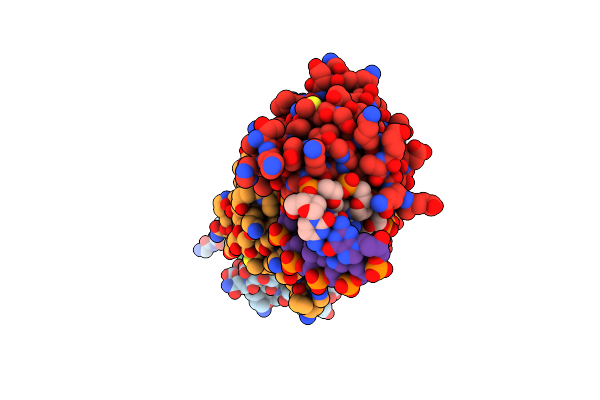
Crystal Structure Of The Human Retinoid X Receptor Dna-Binding Domain Bound To The Human Nr1D1 Response Element
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-02-18 Classification: TRANSCRIPTION/DNA Ligands: ZN, CL, K, NO3 |
 |
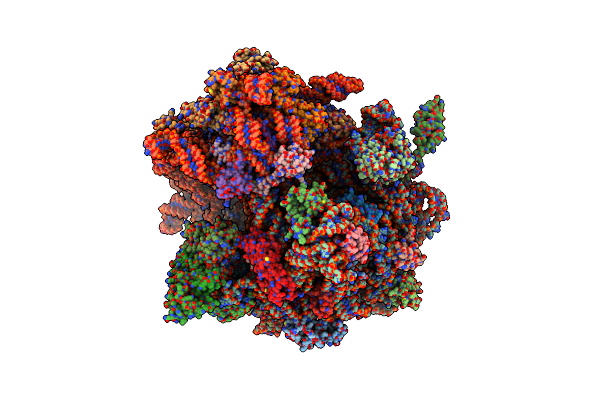
Crystal Structure Of The Human Retinoid X Receptor Dna-Binding Domain Bound To An Idealized Dr1 Response Element
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2015-02-18 Classification: TRANSCRIPTION/DNA Ligands: ZN, CL |
 |
Organism: Thermus thermophilus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Crystal Structure Of The 70S Thermus Thermophilus Ribosome Showing How The 16S 3'-End Mimicks Mrna E And P Codons.
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:5.00 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Crystal Structure Of The 70S Thermus Thermophilus Ribosome With Translocated And Rotated Shine-Dalgarno Duplex.
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
70S Thermus Thermophilous Ribosome Functional Complex With Mrna And E- And P-Site Trnas At 4.5A.
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.51 Å Release Date: 2014-07-09 Classification: RIBOSOME |
 |
The Cryo-Em Structure Of The Palindromic Dna-Bound Usp-Ecr Nuclear Receptor Reveals An Asymmetric Organization With Allosteric Domain Positioning
Organism: Heliothis virescens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:11.60 Å Release Date: 2014-06-25 Classification: NUCLEAR RECEPTOR Ligands: EPH, P1A |
 |
The Nf-Y Transcription Factor Is Structurally And Functionally A Sequence Specific Histone
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2013-01-30 Classification: TRANSCRIPTION/DNA |

