Search Count: 28
 |
Small Subunit Of Yeast Mitochondrial Ribosome In Complex With Mettl17/Rsm22.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: RIBOSOME Ligands: MG, K, LMT, ATP, SF4 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: RIBOSOME Ligands: MG, K, LMT, ATP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: RIBOSOME Ligands: MG, K, LMT, ATP |
 |
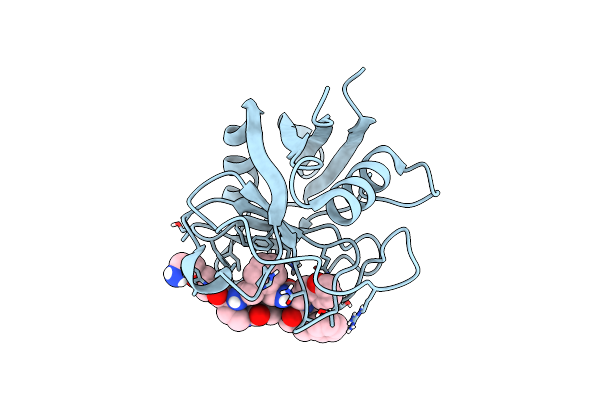
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor Jombt
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I3Q |
 |
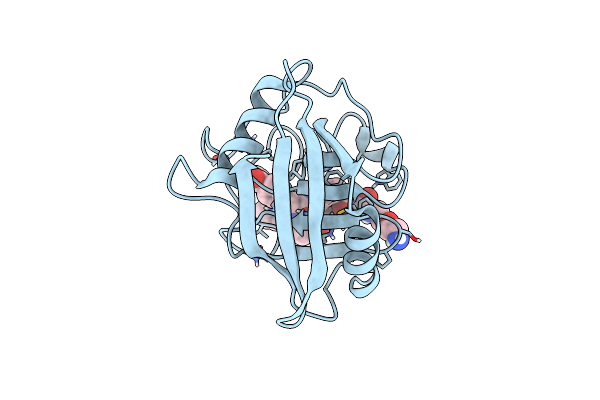
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor A26
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I4F |
 |
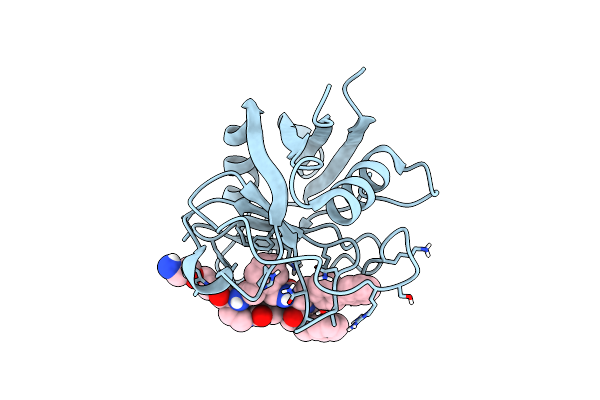
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2022-08-24 Classification: ISOMERASE/ISOMERASE inhibitor Ligands: I44 |
 |
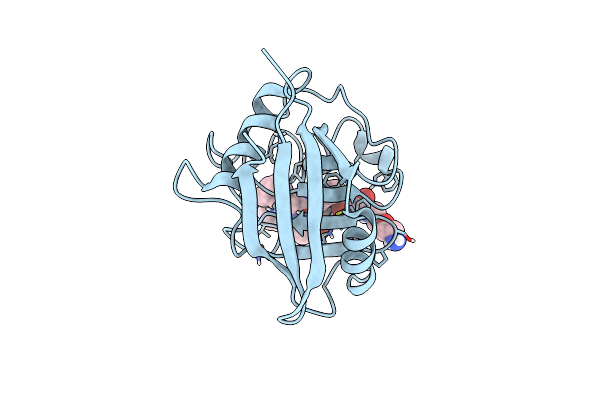
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-08-24 Classification: ISOMERASE/ISOMERASE inhibitor Ligands: I3X |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I5J |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B21
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I56 |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B23
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I5E |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I4Z |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B52
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: I4V |
 |
Structure Of Cyclophilin D Peptidyl-Prolyl Isomerase Domain Bound To Macrocyclic Inhibitor B53
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-08-24 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: I4R |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-09-09 Classification: TRANSPORT PROTEIN |
 |
Double-Stranded Dna-Specific Cytidine Deaminase Type Vi Secretion System Effector And Cognate Immunity Complex From Burkholderia Cenocepacia
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-07-15 Classification: TOXIN Ligands: ZN |
 |
Structure Of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase With Adenosyl Cobalamin
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-13 Classification: ISOMERASE Ligands: B12, 5AD |
 |
Structure Of Mycobacterium Tuberculosis Methylmalonyl-Coa Mutase With Adenosyl Cobalamin
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-13 Classification: ISOMERASE Ligands: B12, 5AD, NJS, K |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-13 Classification: METAL BINDING PROTEIN Ligands: GOL |
 |
Cryo-Em Structure Of The Mitochondrial Calcium Uniporter From N. Fischeri At 3.8 Angstrom Resolution
Organism: Aspergillus fischeri
Method: ELECTRON MICROSCOPY Release Date: 2018-07-11 Classification: TRANSPORT PROTEIN Ligands: CA |

