Search Count: 38
 |
Broad Substrate Scope C-C Oxidation In Cyclodipeptides Catalysed By A Flavin-Dependent Filament
Organism: Nocardiopsis dassonvillei
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: SIGNALING PROTEIN Ligands: OG0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-15 Classification: SIGNALING PROTEIN Ligands: OGR |
 |
Ternary Structure Of Intramolecular Bivalent Glue Degrader Ibg1 Bound To Brd4 And Dcaf16:Ddb1Deltabpb
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: TRANSCRIPTION Ligands: ZN, U79 |
 |
Organism: Thioalkalivibrio sulfidiphilus hl-ebgr7, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: IMMUNE SYSTEM |
 |
Organism: Thioalkalivibrio sulfidiphilus hl-ebgr7
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: GENE REGULATION |
 |
Crystal Structure Of Endonuclease Q Complex With 27-Mer Duplex Substrate With Du At The Active Site
Organism: Pyrococcus furiosus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-03-17 Classification: HYDROLASE/DNA Ligands: ZN, MG, EDO |
 |
Crystal Structure Of Endonuclease Q Complex With 27-Mer Duplex Substrate With Di At The Active Site
Organism: Pyrococcus furiosus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2021-03-17 Classification: HYDROLASE/DNA Ligands: EDO, ZN, MG, CL |
 |
Crystal Structure Of Endonuclease Q Complex With 27-Mer Duplex Substrate With An Abasic Lesion At The Active Site
Organism: Pyrococcus furiosus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2021-03-17 Classification: HYDROLASE/DNA Ligands: ZN, MG |
 |
Crystal Structure Of Endonuclease Q Complex With 27-Mer Duplex Substrate With An Abasic Lesion At The Active Site
Organism: Pyrococcus furiosus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2021-03-17 Classification: HYDROLASE/DNA Ligands: ZN, MG |
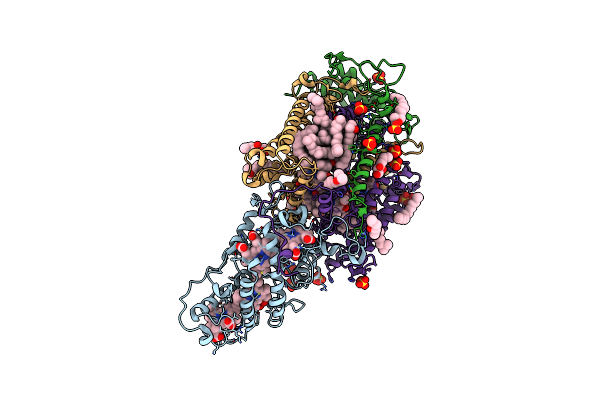 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
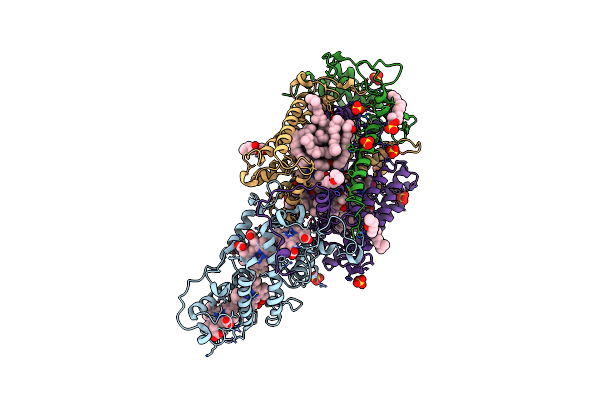 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
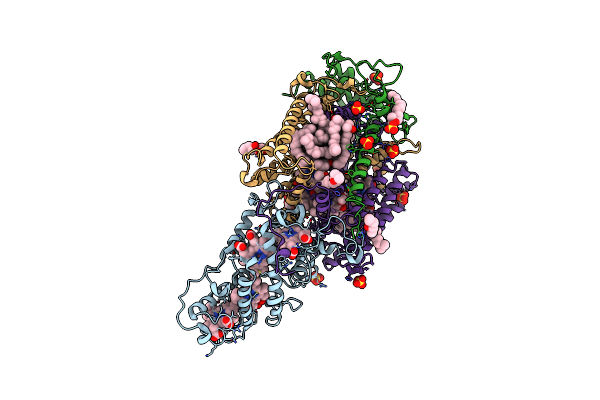 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
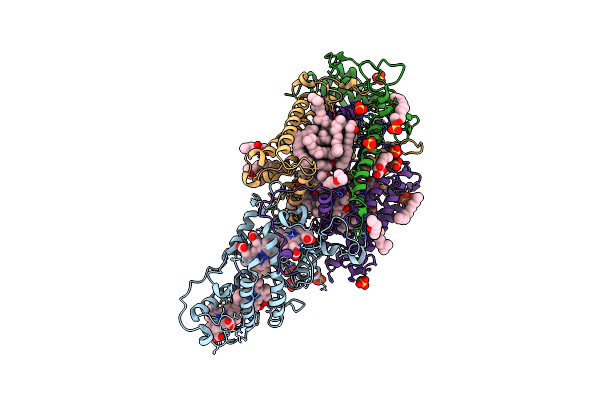 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Structure Of Two Molecules Of The Chromatin Remodelling Enzyme Chd1 Bound To A Nucleosome
Organism: Xenopus laevis, Saccharomyces cerevisiae s288c, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-08-22 Classification: MOTOR PROTEIN Ligands: ADP, BEF |
 |
Structure Of The Chromatin Remodelling Enzyme Chd1 Bound To A Ubiquitinylated Nucleosome
Organism: Petromyzon marinus, Xenopus laevis, Xenopus tropicalis, Homo sapiens, Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:4.50 Å Release Date: 2018-08-08 Classification: MOTOR PROTEIN Ligands: BEF, ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-04-18 Classification: LIGASE Ligands: ZN |

