Search Count: 20
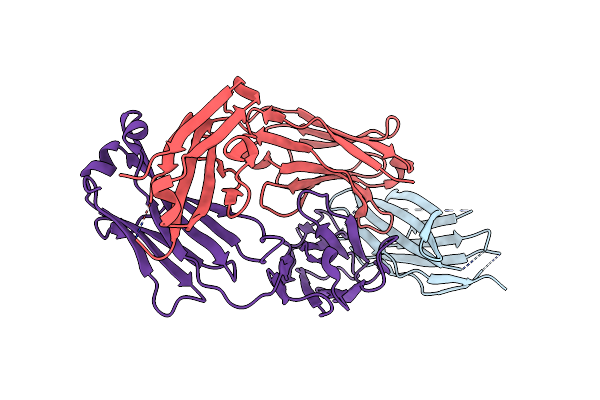 |
Crystal Structure Of Engineered Ipilimumab (Mipi.4) Fab In Complex With Human Ctla-4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-02-05 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
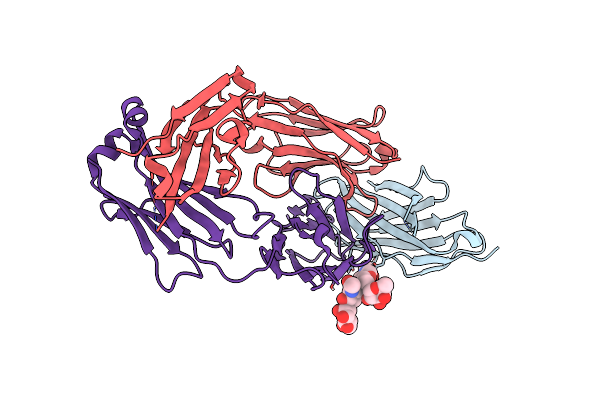 |
Crystal Structure Of Engineered Ipilimumab (Mipi.4) Fab In Complex With Mouse Ctla-4
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-02-05 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
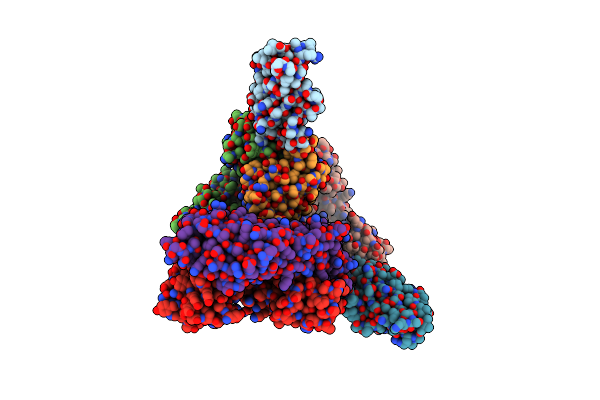 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-02-05 Classification: ANTITUMOR PROTEIN/IMMUNE SYSTEM |
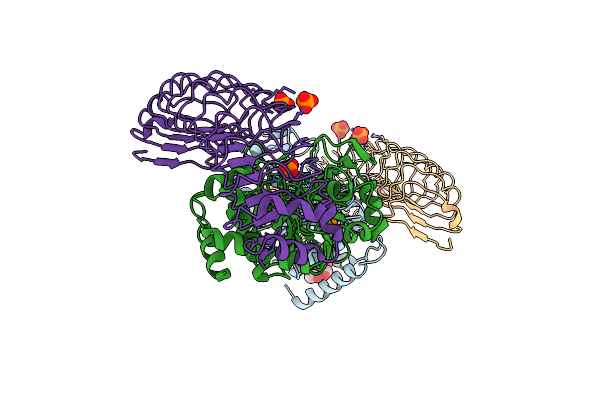 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: PO4, FE, CL, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: PO4, MN, CL |
 |
Organism: Homo sapiens, Microcystis aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2019-09-18 Classification: HYDROLASE/TOXIN Ligands: MN |
 |
Organism: Homo sapiens, Microcystis aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2019-09-18 Classification: HYDROLASE/TOXIN Ligands: MN, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: MN, PO4, GOL |
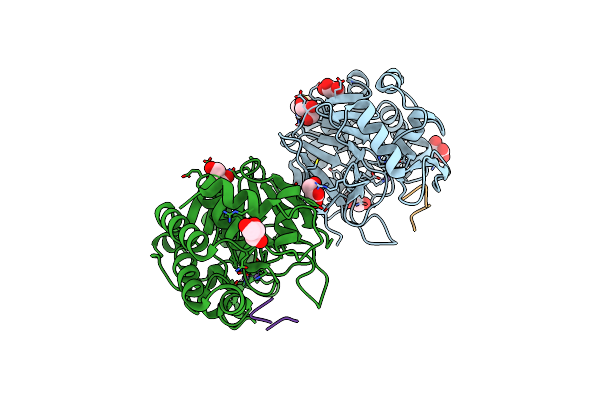 |
Organism: Homo sapiens, Microcystis aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2019-09-18 Classification: HYDROLASE/TOXIN Ligands: MN, GOL, DMS, CL |
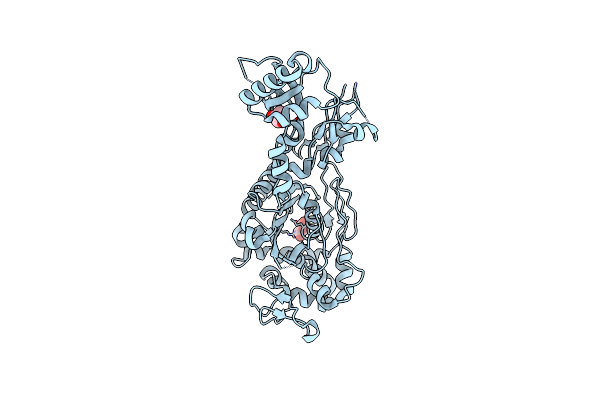 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: GOL, CL |
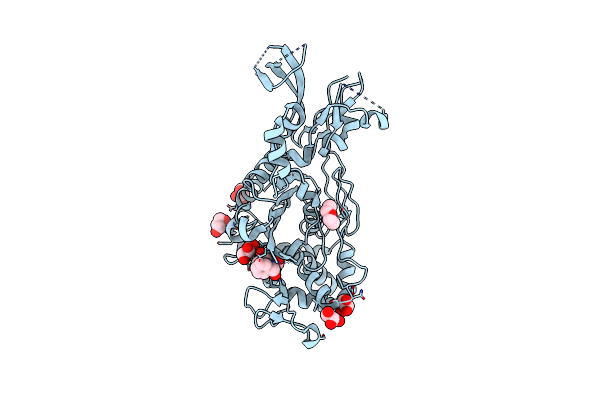 |
Crystal Structure Of Penicillin-Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Benzylpenicillin Bound Form.
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: GOL, CL, PEG, PNM |
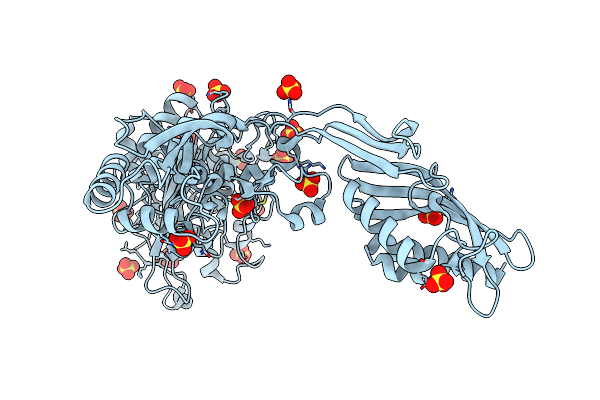 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Open Conformation
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-31 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Imipenem-Bound Form
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: IM2, SO4 |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Benzylpenicilin-Bound Form
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: PNM, SO4 |
 |
Crystal Structure Of Pencillin Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Imipenem-Bound Form
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: IM2, PO4 |
 |
Crystal Structure Of Penicillin-Binding Protein 4 (Pbp4) From Enterococcus Faecalis In The Ceftaroline-Bound Form
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2018-10-31 Classification: protein binding/antibiotic Ligands: AI8, PO4, GOL |
 |
Crystal Structure Of Penicillin Binding Protein 5 (Pbp5) From Enterococcus Faecium In The Closed Conformation
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2018-10-31 Classification: PROTEIN BINDING |
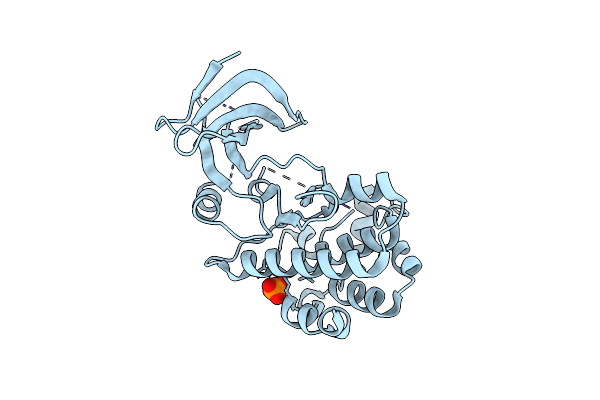 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: BR |
 |
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2012-05-23 Classification: TRANSFERASE |

