Search Count: 41
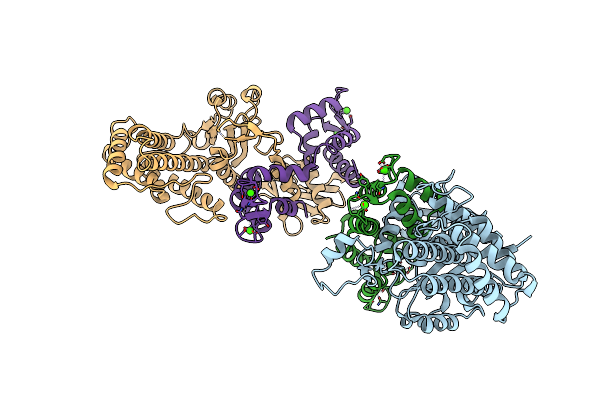 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: CA |
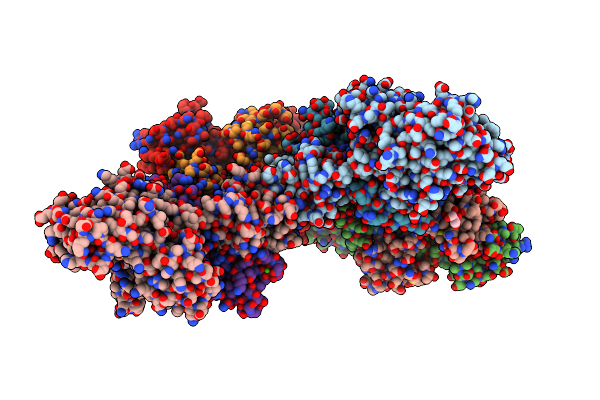 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin And A Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: NAD, CA, GOL |
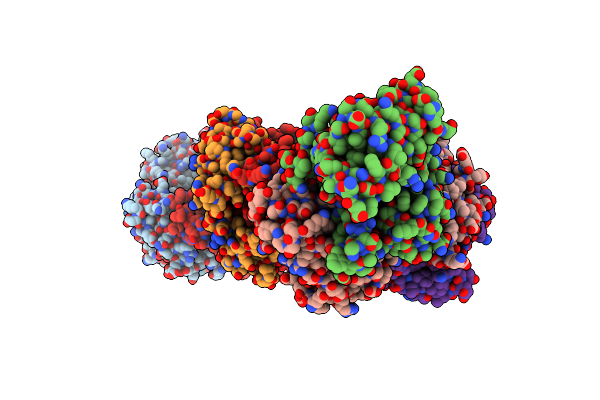 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Free Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: MG |
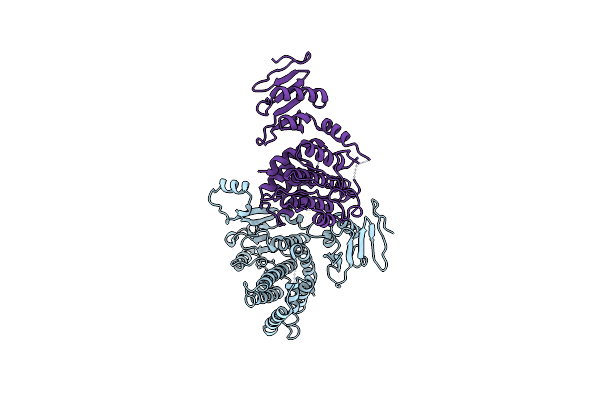 |
Crystal Structure Of The Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) From Vibrio Vulnificus
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2024-07-10 Classification: TOXIN |
 |
Crystal Structure At 1.3 Angstroms Resolution Of A Novel Udg, Udgx, From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, PO4, URA |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: DU, SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
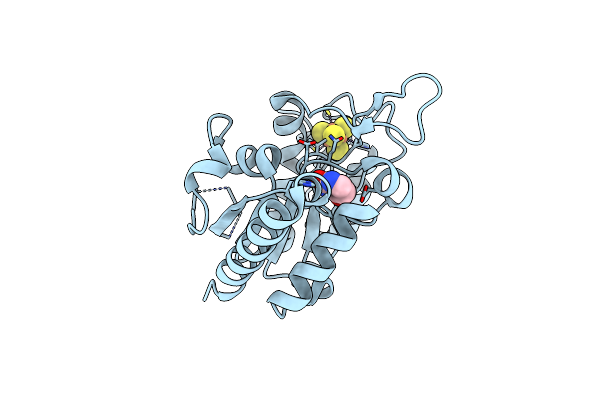 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |
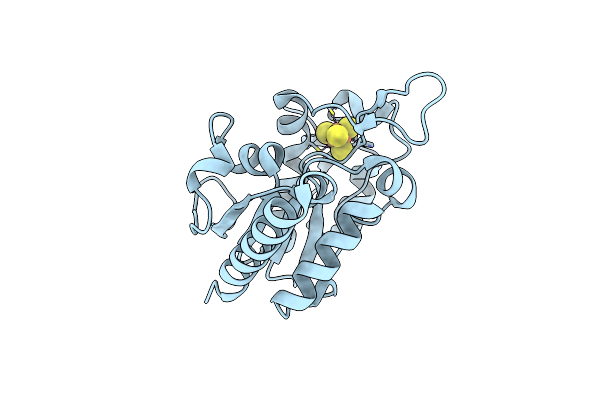 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
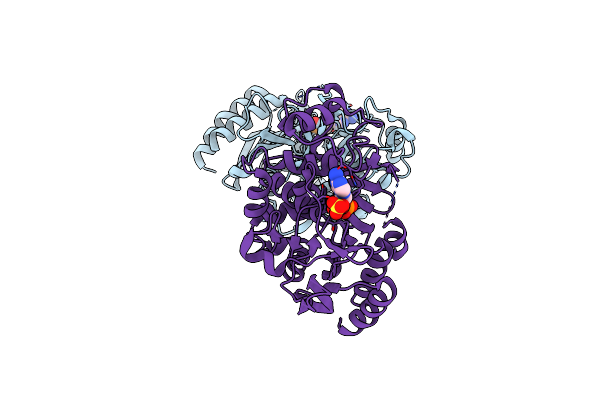 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-10-11 Classification: HYDROLASE Ligands: AMP, SO4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-06-07 Classification: OXIDOREDUCTASE |
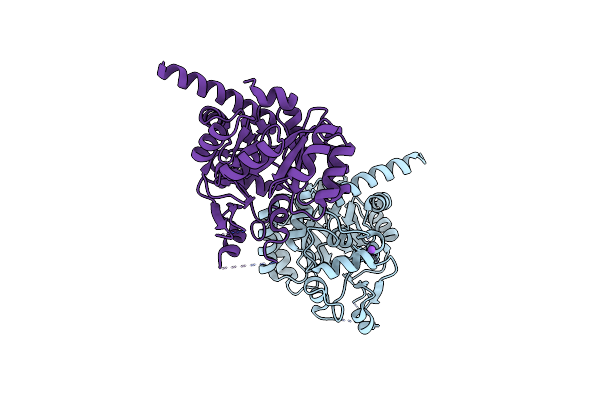 |
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE |
 |
Organism: Enterobacter aerogenes
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-12-07 Classification: HYDROLASE Ligands: CD, AMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-12-07 Classification: HYDROLASE Ligands: AMP, SO4, EDO |
 |
Crystal Structure Of A Response Regulator Spr1814 From Streptococcus Pneumoniae Reveals Unique Interdomain Contacts Among Narl Family Proteins
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-06-05 Classification: TRANSCRIPTION ACTIVATOR |
 |
Crystal Structure Of Receiver Domain Of Putative Narl Family Response Regulator Spr1814 From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-23 Classification: TRANSCRIPTION REGULATOR Ligands: MG |
 |
Crystal Structure Of Receiver Domain Of Putative Narl Family Response Regulator Spr1814 From Streptococcus Pneumoniae In The Presence Of The Phosphoryl Analog Beryllofluoride
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2012-05-23 Classification: TRANSCRIPTION REGULATOR Ligands: MG, BEF |
 |
Crystal Structure Of A Glutathione S-Transferase From Antarctic Clam Laternula Elliptica
Organism: Laternula elliptica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-01 Classification: TRANSFERASE |
 |
Crystal Structure Of A Glutathione-S-Transferase From Antarctic Clam Laternula Elliptica In A Complex With Glutathione
Organism: Laternula elliptica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-02-01 Classification: TRANSFERASE Ligands: GSH |

