Search Count: 95
 |
Crystal Structure Of Apo-Form Artificial Metalloprotein Incorporating A Tp Ligand
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of Cu-Bound Artificial Metalloprotein Incorporating A Tp Ligand
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2025-10-15 Classification: METAL BINDING PROTEIN Ligands: CU, NA, SO4 |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: CA |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Bound Calmodulin And A Nicotinamide Adenine Dinucleotide (Nad+)
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: NAD, CA, GOL |
 |
Crystal Structure Of Vibrio Vulnificus Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) Complexed With Ca2+-Free Calmodulin
Organism: Vibrio vulnificus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2024-07-10 Classification: TOXIN Ligands: MG |
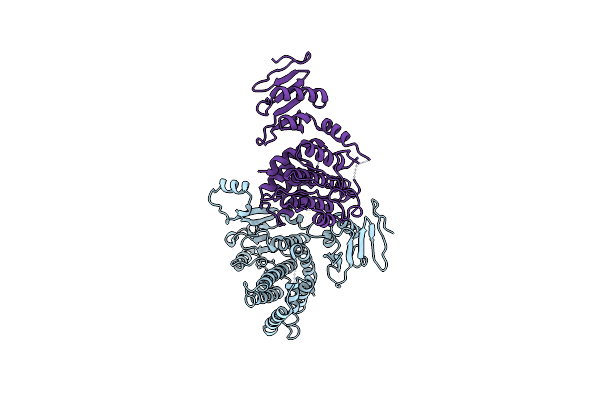 |
Crystal Structure Of The Rid-Dependent Transforming Nadase Domain (Rdtnd)/Calmodulin-Binding Domain Of Rho Inactivation Domain (Rid-Cbd) From Vibrio Vulnificus
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2024-07-10 Classification: TOXIN |
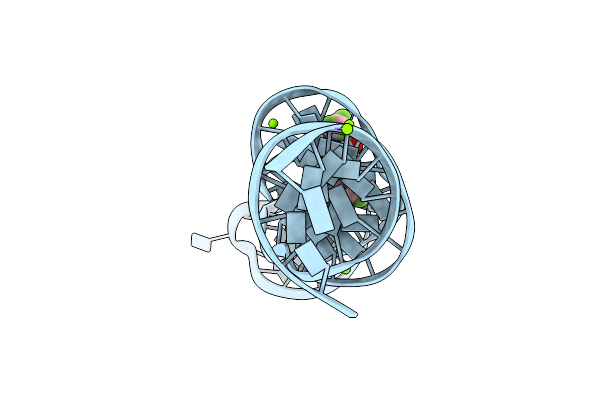 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-05-10 Classification: DNA Ligands: TL, MG, 2ZY |
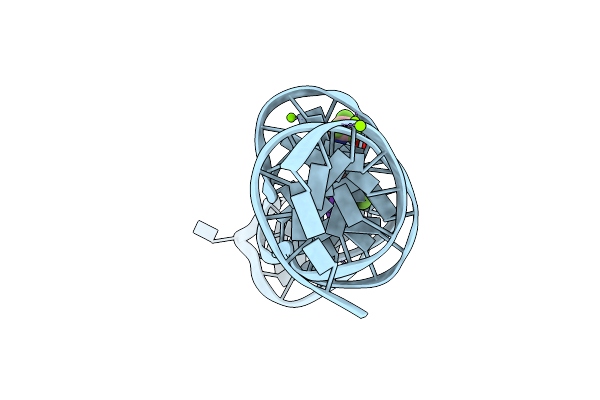 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, 2ZY |
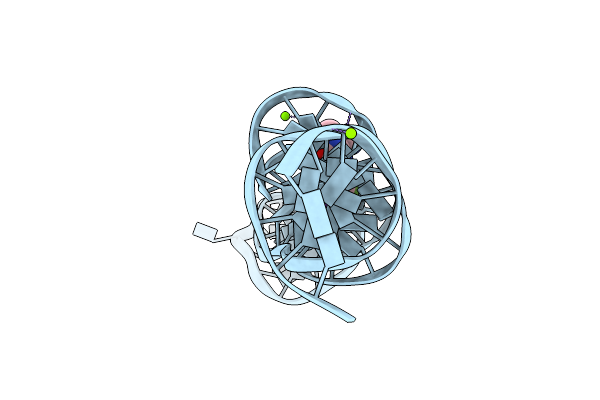 |
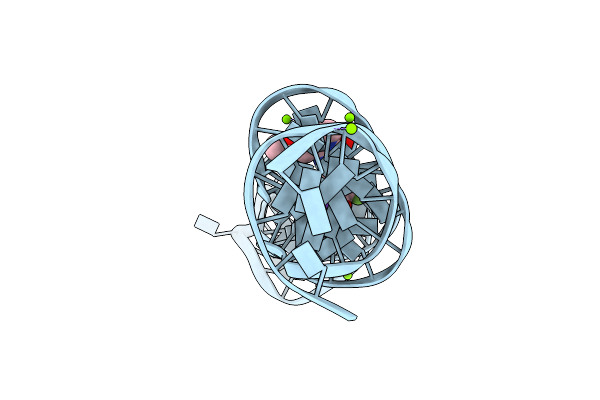
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, 747 |
 |
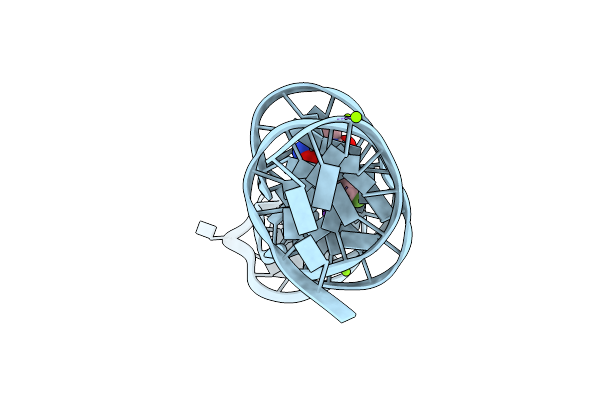
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, X5R |
 |
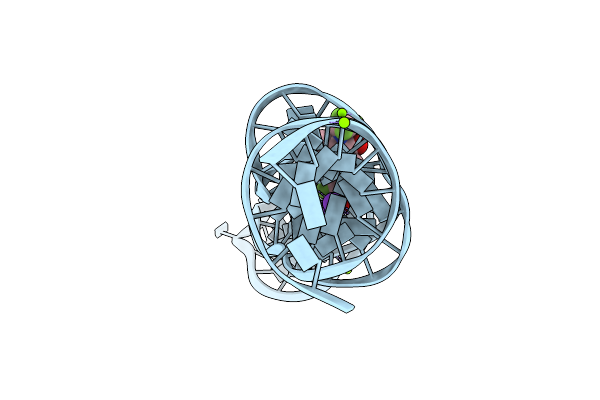
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, 747 |
 |
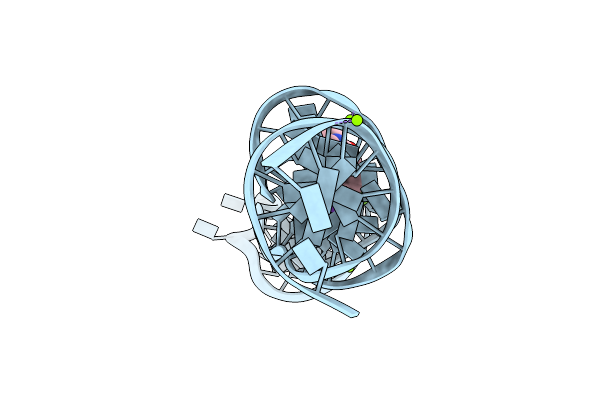
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, 2ZY |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, 747 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-05-10 Classification: DNA Ligands: K, MG, X5R |
 |
Organism: Homo sapiens, Human gammaherpesvirus 4
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-12 Classification: IMMUNE SYSTEM Ligands: CL, GOL |
 |
Crystal Structure At 1.3 Angstroms Resolution Of A Novel Udg, Udgx, From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, PO4, URA |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: DU, SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Mycobacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2019-05-29 Classification: DNA BINDING PROTEIN Ligands: SF4, URA |

