Search Count: 22
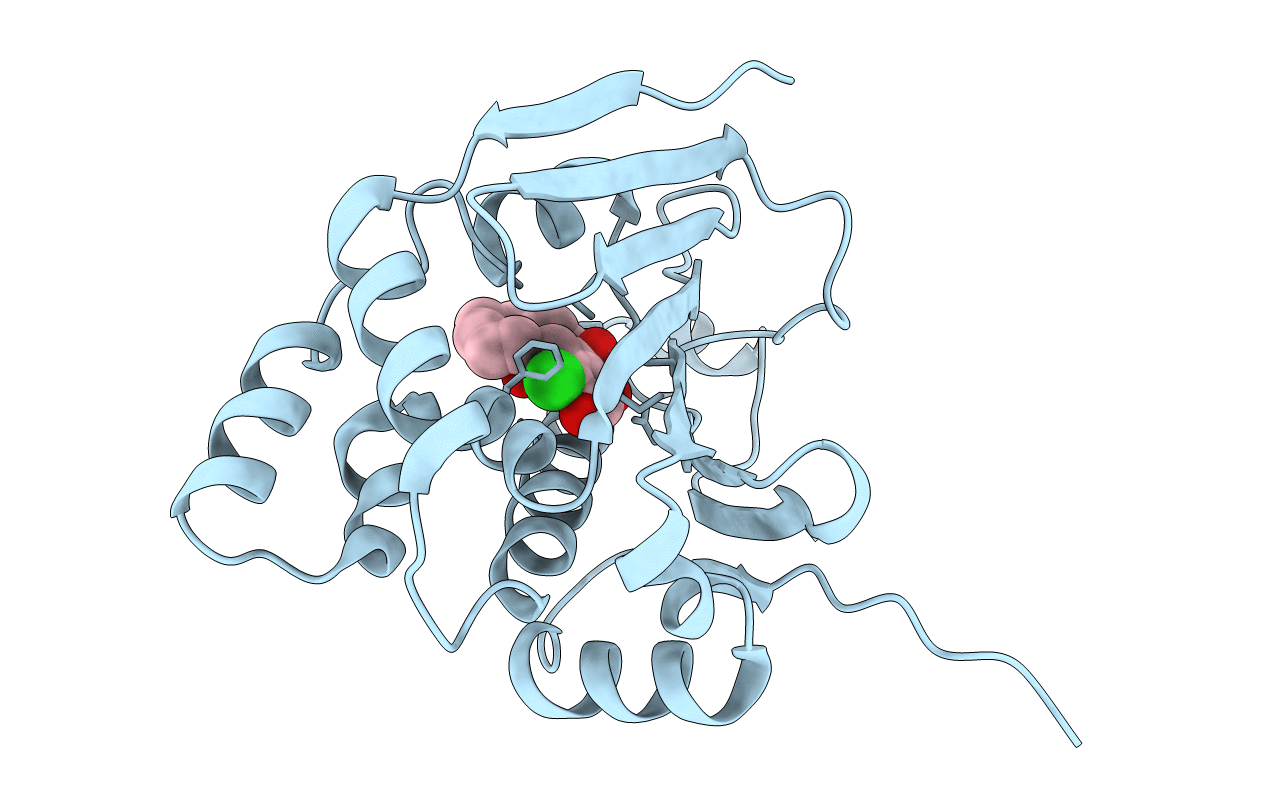 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2014-01-29 Classification: CHAPERONE Ligands: 7FK |
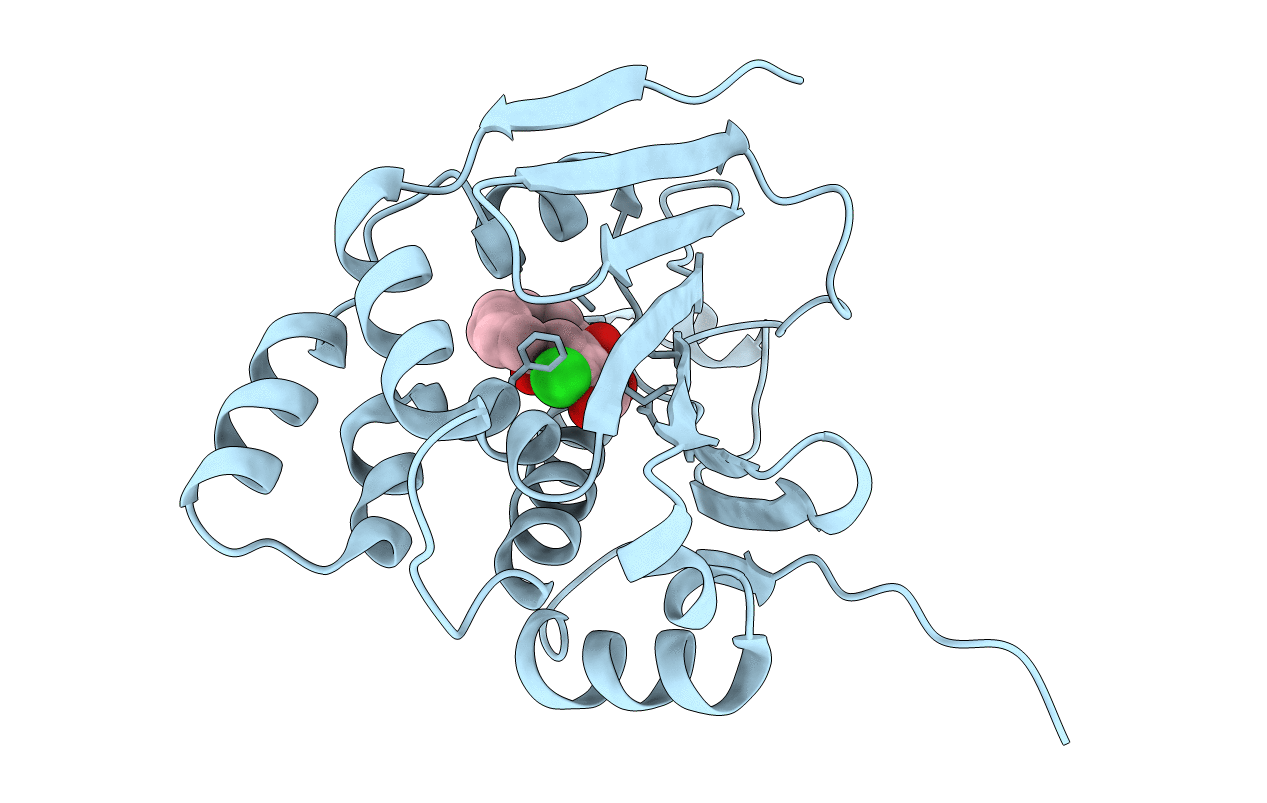 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2014-01-29 Classification: CHAPERONE Ligands: BO5 |
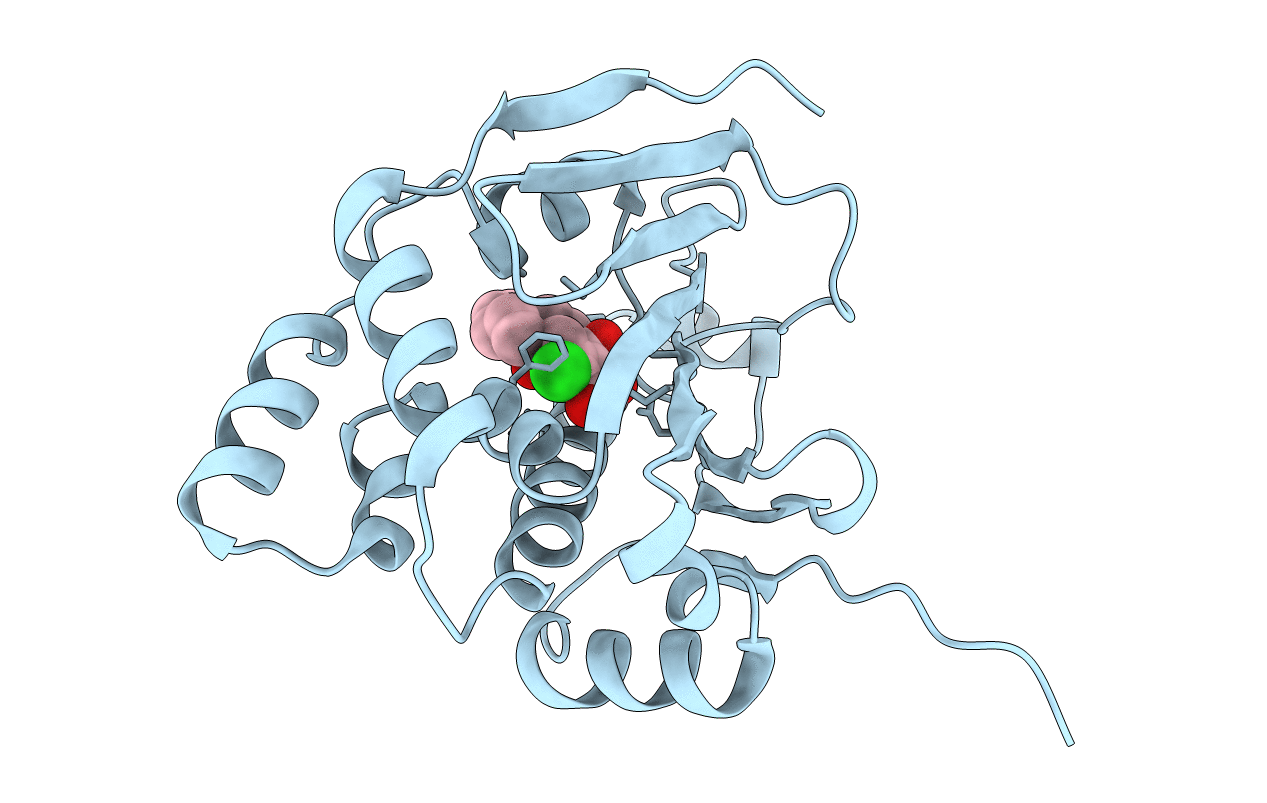 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2014-01-29 Classification: CHAPERONE Ligands: L4V |
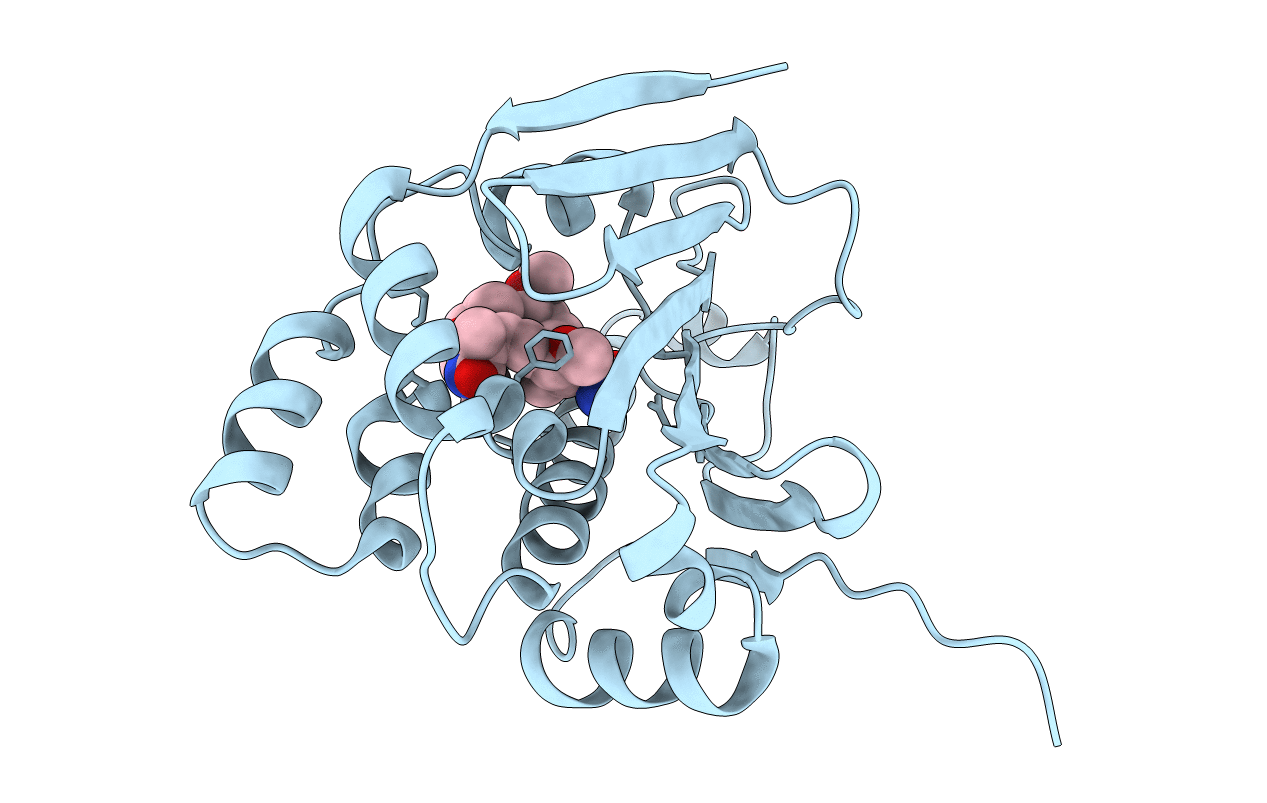 |
The Structure Of Modified Benzoquinone Ansamycins Bound To Yeast N- Terminal Hsp90
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-04-03 Classification: CHAPERONE Ligands: 4QS |
 |
The Structure Of Modified Benzoquinone Ansamycins Bound To Yeast N- Terminal Hsp90
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-04-03 Classification: CHAPERONE Ligands: 59C, CL |
 |
The Structure Of Modified Benzoquinone Ansamycins Bound To Yeast N- Terminal Hsp90
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2013-04-03 Classification: CHAPERONE Ligands: 8TO |
 |
The Structure Of Modified Benzoquinone Ansamycins Bound To Yeast N- Terminal Hsp90
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-04-03 Classification: CHAPERONE Ligands: 62U, NI |
 |
The Structure Of Modified Benzoquinone Ansamycins Bound To Yeast N- Terminal Hsp90
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-03 Classification: CHAPERONE Ligands: 814, NI |
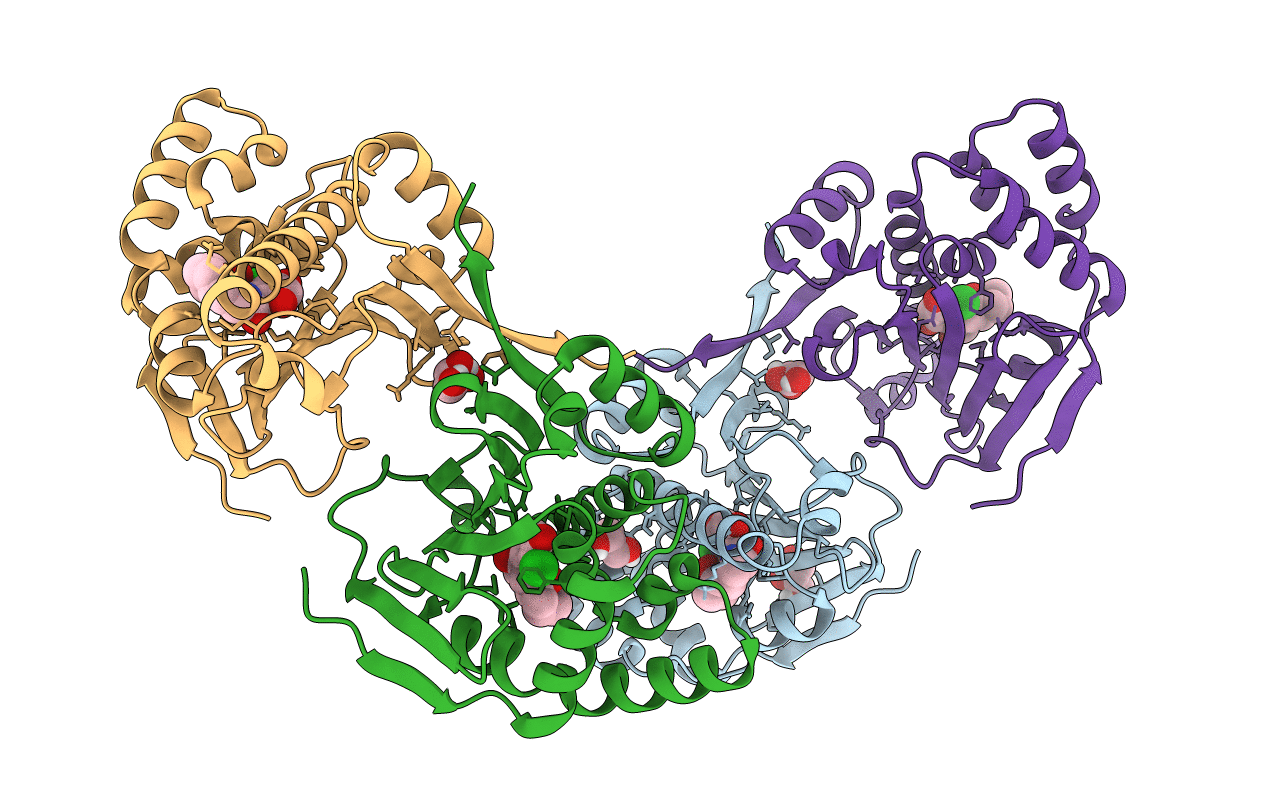 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-11-16 Classification: CHAPERONE Ligands: 13C, GOL |
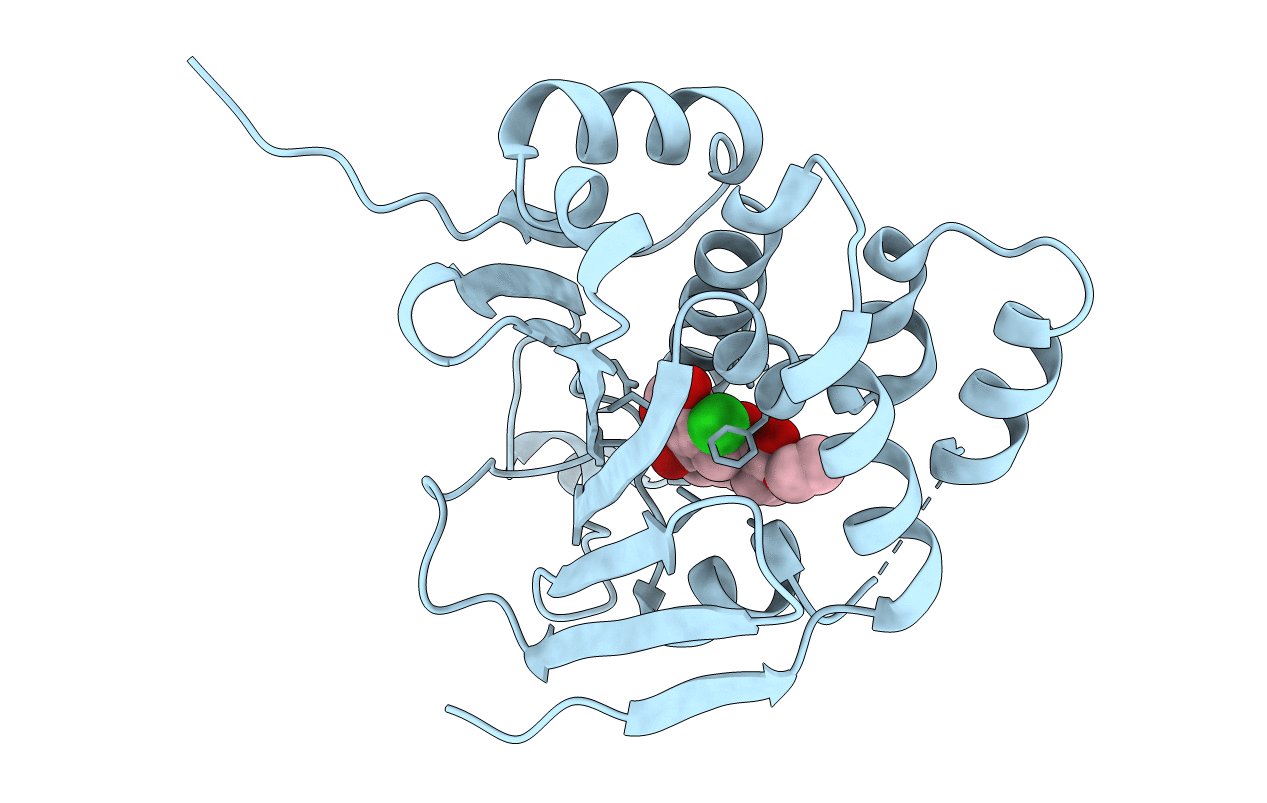 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-11-16 Classification: CHAPERONE Ligands: 13I |
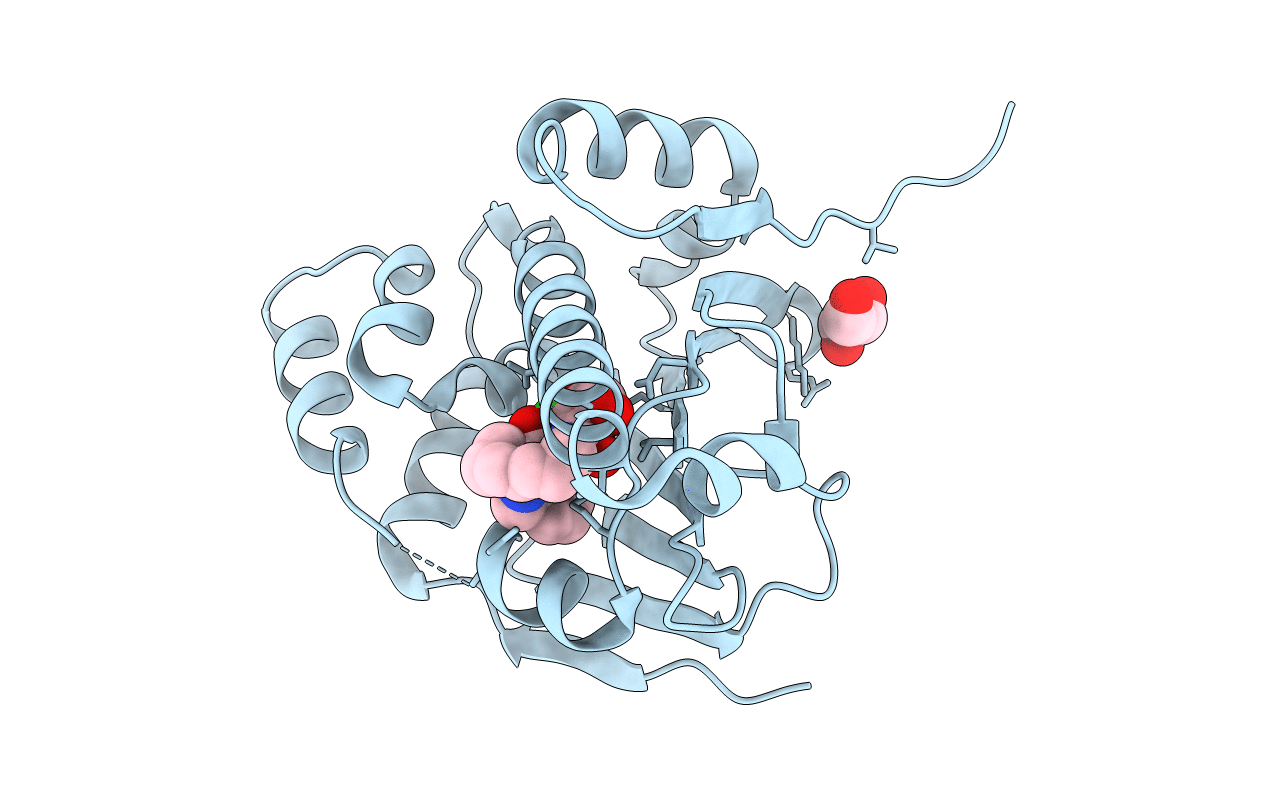 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-11-16 Classification: CHAPERONE Ligands: 13N, GOL |
 |
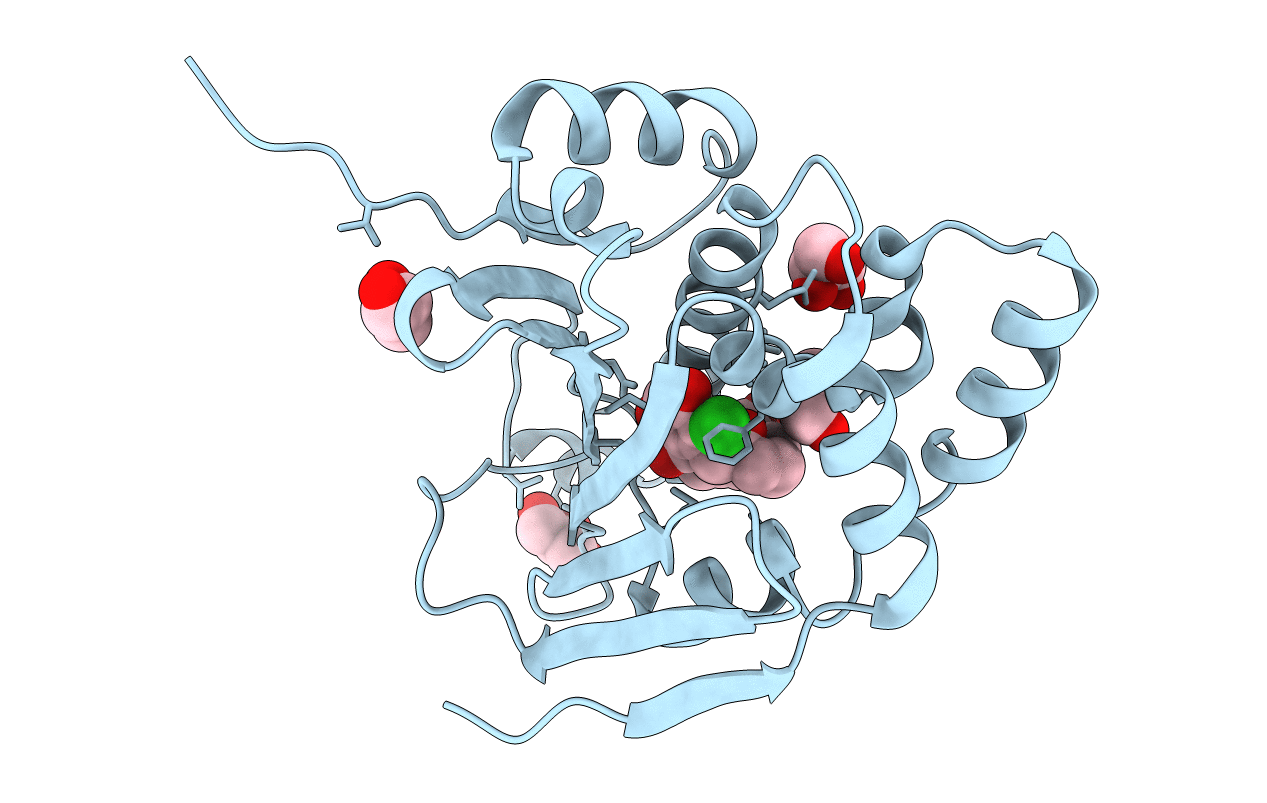
Crystal Structure Of Quinone Reductase 2 In Complex With The Indolequinone Mac627
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-05-11 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: ZN, FAD, O73 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-08-11 Classification: CHAPERONE Ligands: XD6, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-11-30 Classification: CHAPERONE Ligands: NP4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-11-30 Classification: CHAPERONE Ligands: NP5 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-11-30 Classification: CHAPERONE Ligands: M1S |
 |
A Radicicol Analogue Bound To The Atp Binding Site Of The N-Terminal Domain Of The Yeast Hsp90 Chaperone
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-11-29 Classification: CHAPERONE Ligands: P2N |
 |
Complex Of Human Recombinant Nad(P)H:Quinone Oxide Reductase Type 1 With 5-Methoxy-1,2-Dimethyl-3-(Phenoxymethyl)Indole-4,7-Dione (Es1340)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-01-16 Classification: OXIDOREDUCTASE Ligands: 340, FAD |
 |
Complex Of Human Nad(P)H Quinone Oxidoreductase With 5-Methoxy-1,2-Dimethyl-3-(4-Nitrophenoxymethyl)Indole-4,7-Dione (Es936)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-01-16 Classification: OXIDOREDUCTASE Ligands: FAD, 936 |
 |
Crystal Structure Of A Complex Of Human Nad[P]H-Quinone Oxidoreductase And A Chemotherapeutic Drug (E09) At 2.5 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-09-12 Classification: OXIDOREDUCTASE Ligands: FAD, E09 |

