Search Count: 36
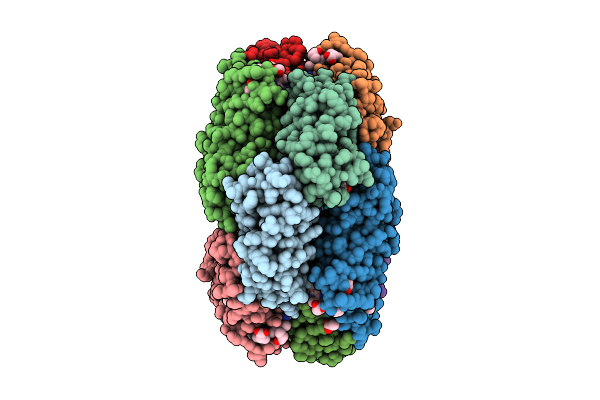 |
X-Ray Structure Of The Adduct Formed By Dirhodium Tetraacetate With A C-Phycocyanin
Organism: Galdieria phlegrea
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: PHOTOSYNTHESIS Ligands: CYC, A1JTN, ACT, DOD |
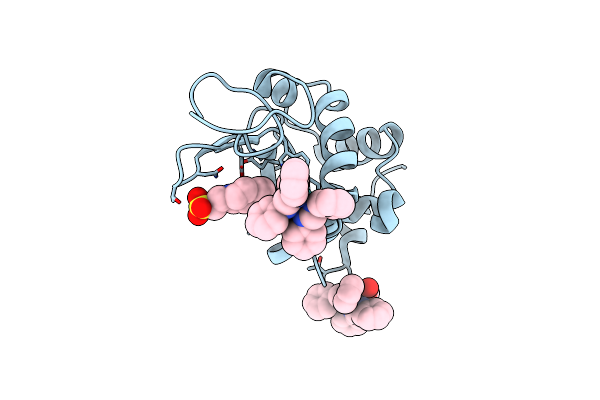 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of Lysozyme With [Ru2Cl(Dphf)2(O2Cch3)2] In Condition B
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2023-09-13 Classification: PROTEIN BINDING Ligands: EPE, ZJK |
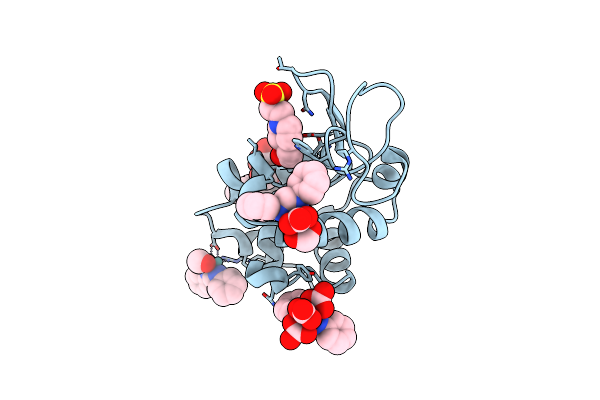 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of Lysozyme With K2[Ru2(Dphf)(Co3)3] In Condition B
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2023-09-13 Classification: PROTEIN BINDING Ligands: EPE, ZWL, YWV, ZWO, ZXE |
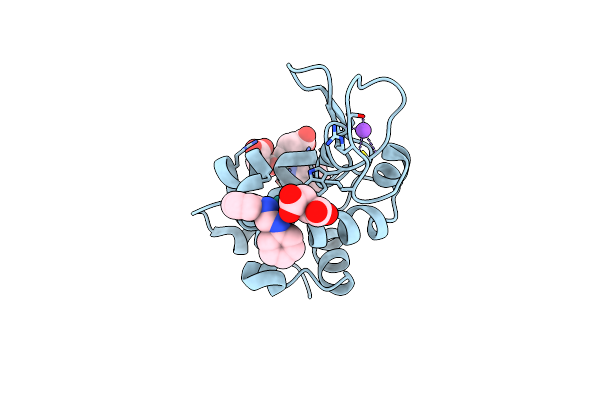 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of Lysozyme With [Ru2Cl(Dphf)(O2Cch3)3] In Condition A
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2023-09-13 Classification: PROTEIN BINDING Ligands: ZQ2, SIN, ZWH, NA |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of Lysozyme With [Ru2Cl(Dphf)2(O2Cch3)2] In Condition A
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2023-09-13 Classification: PROTEIN BINDING |
 |
Limonene Epoxide Low Ph Soak Of Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: LEO, CL, EDO |
 |
Cyclohexane Epoxide Low Ph Soak Of Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: 7EX, PEG, EDO |
 |
Halogenated Product Of Limonene Epoxide Turnover By Epoxide Hydrolase From Metagenomic Source Ch65
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: M1U, CL, EDO, NA |
 |
Cyclohexane Epoxide Soak Of Epoxide Hydrolase From Metagenomic Source Ch65 Resulting In Halogenated Compound In The Active Site
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: 2CH, EDO, PGE, CL, P6G |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Five-Coordinate Pt(Ii) Complex, 1-Me,Me, With Hewl At Ph 7.5
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: EPE, ACT, R0I, DMS |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Five-Coordinate Pt(Ii) Complex, 1-Me,Me, With Hewl At Ph 4.0
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: NO3, R0I, PT |
 |
Organism: Porphyridium purpureum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-02-08 Classification: PHOTOSYNTHESIS Ligands: PEB, SO4 |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2022-12-28 Classification: TRANSPORT PROTEIN Ligands: CD, SO4, AU, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2022-12-28 Classification: TRANSPORT PROTEIN Ligands: CL, MG, FE, AU |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Gold(I) N-Heterocyclic Carbene Complex Au1 With Rnase A
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-07-13 Classification: RNA BINDING PROTEIN Ligands: AU, SO4 |
 |
X-Ray Structure Of The Adduct Formed Upon Reaction Of The Gold(I) N-Heterocyclic Carbene Complex Au2 With Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: DMS, AU |
 |
Cryo-Em Structure Of Mal De Rio Cuarto Virus P9-1 Viroplasm Protein (Decamer)
Organism: Mal de rio cuarto virus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Mal De Rio Cuarto Virus P9-1 Viroplasm Protein (Dodecamer)
Organism: Mal de rio cuarto virus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-15 Classification: VIRAL PROTEIN |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-03-23 Classification: TRANSPORT PROTEIN Ligands: PT |
 |
Crystal Structure Of Mal De Rio Cuarto Virus P9-1 Viroplasm Protein (C-Arm Deletion Mutant)
Organism: Mal de rio cuarto virus
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2021-03-17 Classification: VIRAL PROTEIN |

