Search Count: 153
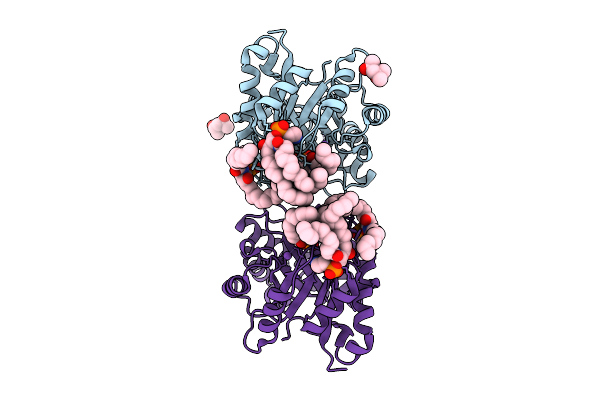 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 2.2 A Resolution From A 2-Day Old Crystal
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
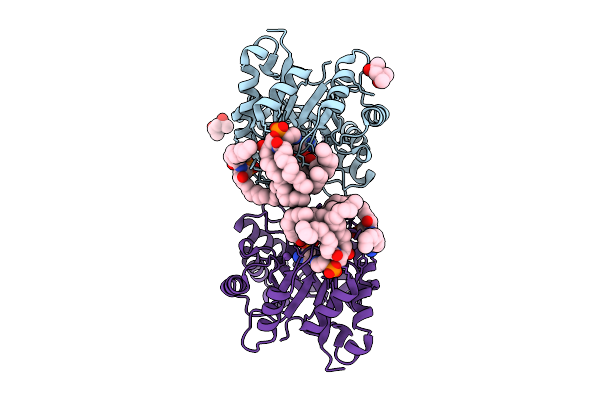 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Substrate Sphingolipids At 2.60 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: LYASE Ligands: A1A43, NA, MG, MPD |
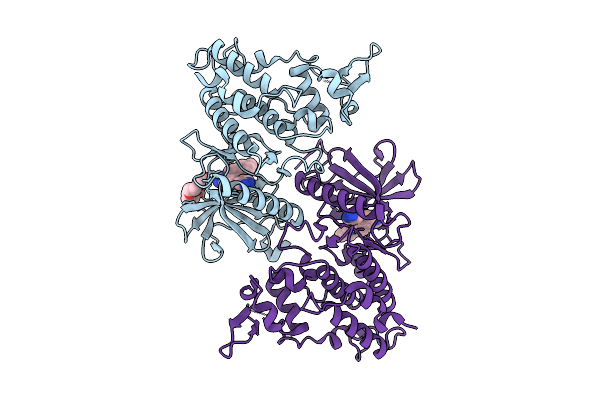 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: A1A04, PG4 |
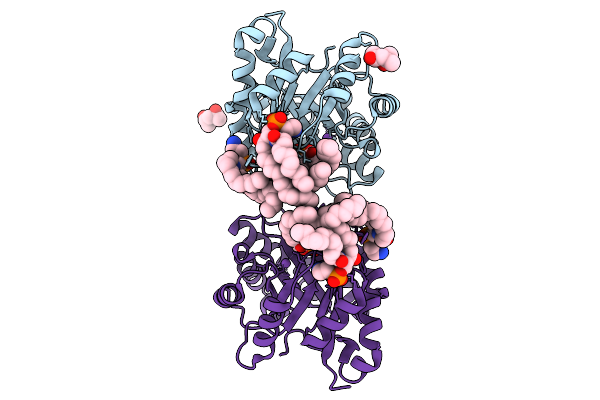 |
Structure Of Phospholipase D Betaib1I From Sicarius Terrosus Venom, H47N Mutant Bound To Product And Substrate Sphingolipids At 1.85 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LYASE Ligands: A1A43, A1A44, NA, MG, MPD |
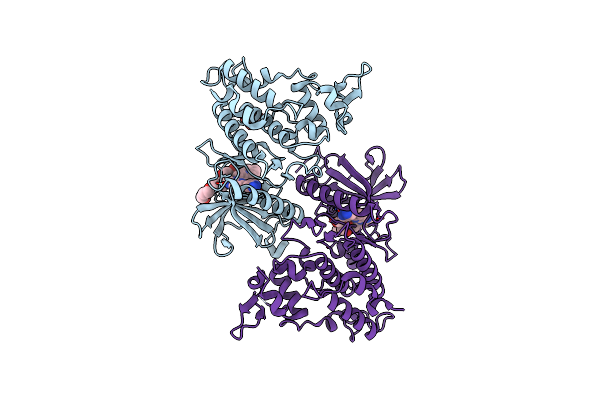 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-15 Classification: LIGASE/LIGASE INHIBITOR Ligands: XIR, PG4, GOL |
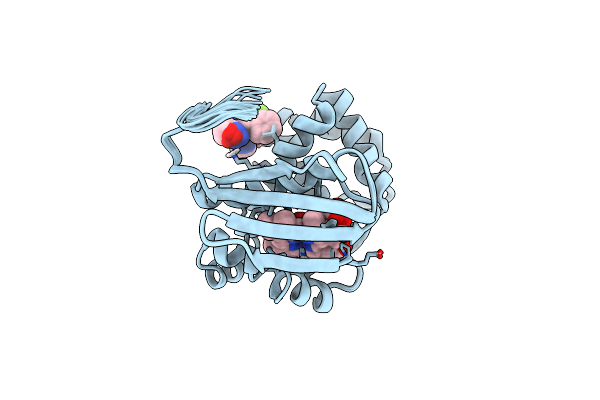 |
Solution Structure Of The Iwp-051-Bound H-Nox From Shewanella Woodyi In The Fe(Ii)Co Ligation State
Organism: Shewanella woodyi
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: SIGNALING PROTEIN Ligands: IWP, HEM, CMO |
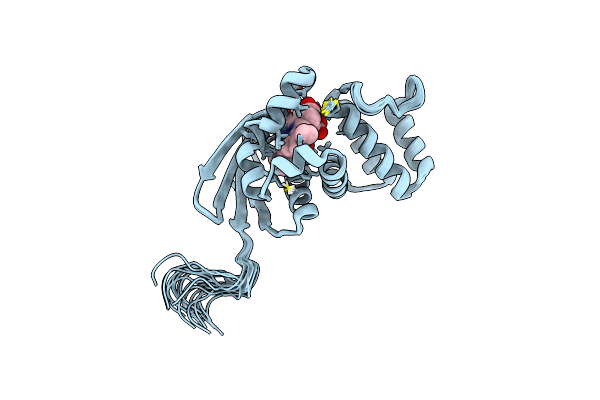 |
Solution Structure Of The H-Nox Protein From Shewanella Woodyi In The Fe(Ii)Co Ligation State
Organism: Shewanella woodyi
Method: SOLUTION NMR Release Date: 2020-05-06 Classification: SIGNALING PROTEIN Ligands: CMO, HEM |
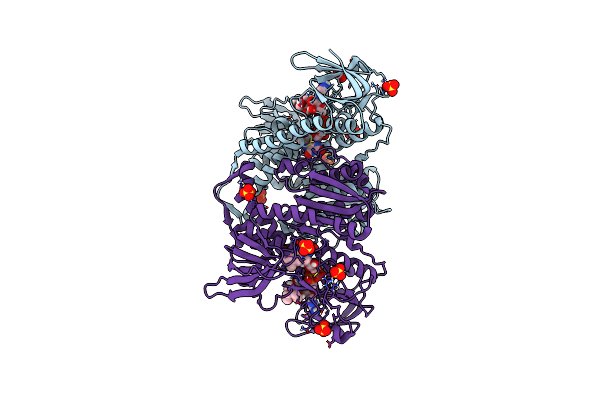 |
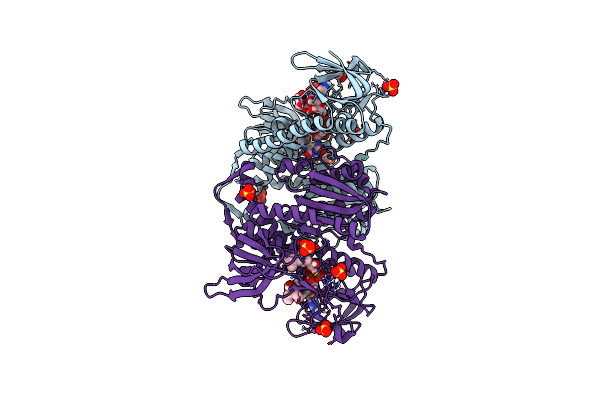
Crystal Structure Of The Disease-Causing G194C Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, BTB |
 |
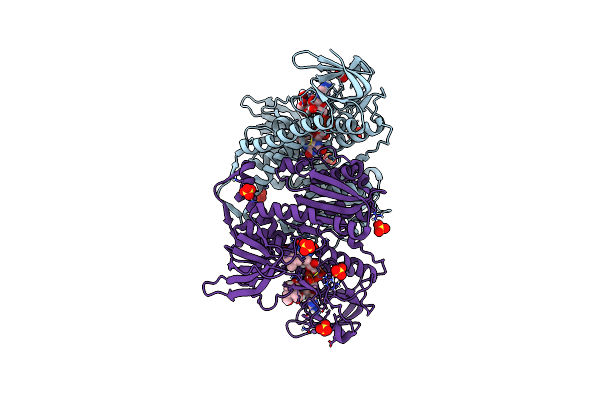
Crystal Structure Of The Human Dihydrolipoamide Dehydrogenase At 1.75 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, BTB, SO4 |
 |
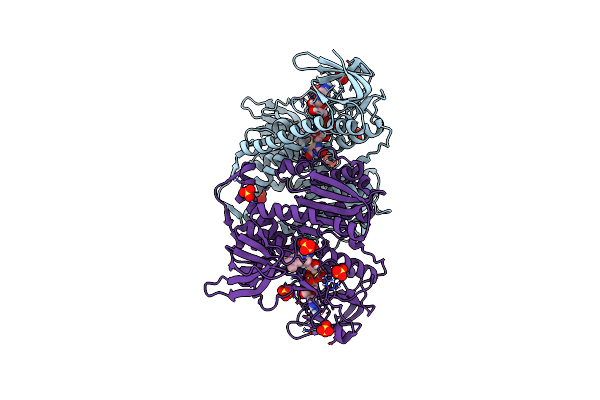
Crystal Structure Of The Disease-Causing R460G Mutant Of The Human Dihydrolipoamide Dehydrogenase At 1.44 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, BTB |
 |
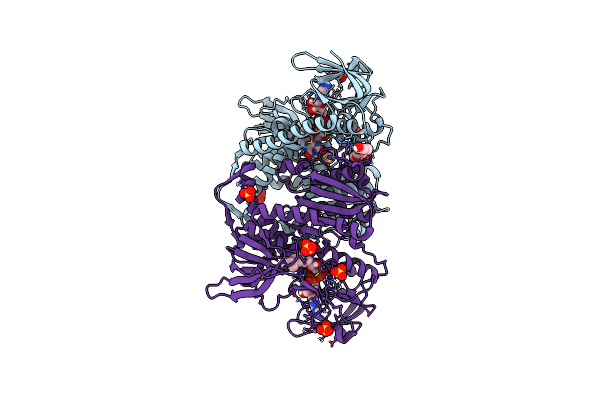
Crystal Structure Of The Disease-Causing R447G Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
Crystal Structure Of The Disease-Causing I445M Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, 1PE |
 |
Crystal Structure Of The Disease-Causing G426E Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, 1PE |
 |
Crystal Structure Of The Disease-Causing P453L Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
Crystal Structure Of The R460G Disease-Causing Mutant Of The Human Dihydrolipoamide Dehydrogenase.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-09-04 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, TRS |
 |
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-11-07 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-07-11 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Organism: Cimex lectularius
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-09-17 Classification: TRANSPORT PROTEIN Ligands: HEM |
 |
Organism: Cimex lectularius
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-09-17 Classification: TRANSPORT PROTEIN Ligands: HEM |
 |
Organism: Cimex lectularius
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-06-04 Classification: TRANSPORT PROTEIN Ligands: HEM, NO |

