Search Count: 1,079
 |
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-02-21 Classification: VIRAL PROTEIN |
 |
Organism: Salmonella enterica
Method: SOLUTION NMR Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In D6-Dmso With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In D6-Dmso With Cis/Trans Switching (A-Cc Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In D6-Dmso With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In 50% D6-Dmso And 50% Water With Cis/Trans Switching (Cc Conformation, 50%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In Cdcl3 With Cis/Trans Switching (A-Tt Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In Cdcl3 With Cis/Trans Switching
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In Cdcl3 With Cis/Trans Switching (Tt Conformation, 47%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2022-09-14 Classification: DE NOVO PROTEIN Ligands: HOH |
 |
Solution Nmr Structure Of 9-Residue Rosetta-Designed Cyclic Peptide D9.16 In Cdcl3 With Cis/Trans Switching (B-Tc Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.31 In D6-Dmso With Cis/Trans Switching (B-Ct Conformation)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In 50% D6-Dmso And 50% Water With Cis/Trans Switching (Cc Conformation, 50%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of 8-Residue Rosetta-Designed Cyclic Peptide D8.21 In Cdcl3 With Cis/Trans Switching (Tc Conformation, 53%)
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-09-14 Classification: DE NOVO PROTEIN |
 |
Solution Nmr Structure Of The Pbs Linker Polypeptide Domain (Fragment 254-400) Of Phycobilisome Linker Protein Apce From Synechocystis Sp. Pcc 6803 Refined With Nh Rdcs. Northeast Structural Genomics Consortium Target Sgr209C
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: SOLUTION NMR Release Date: 2022-08-10 Classification: LYASE |
 |
Solution Nmr Of The Specialized Apo-Acyl Carrier Protein (Rpa2022) From Rhodopseudomonas Palustris, Refined Without Rdcs. Northeast Structural Genomics Consortium Target Rpr324
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: SOLUTION NMR Release Date: 2022-08-10 Classification: TRANSFERASE |
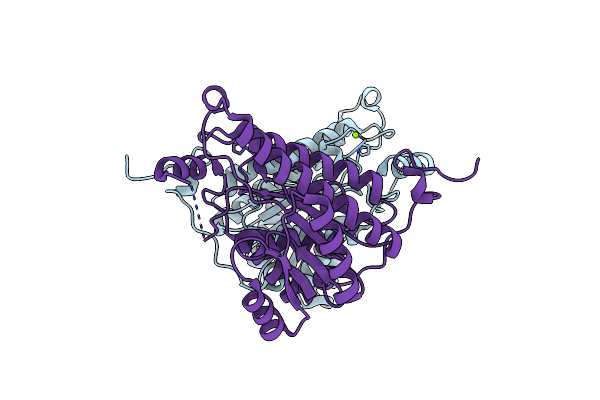 |
Crystal Structures Of (S)-Carbonyl Reductases From Candida Parapsilosis In Different Oligomerization States
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: MG |
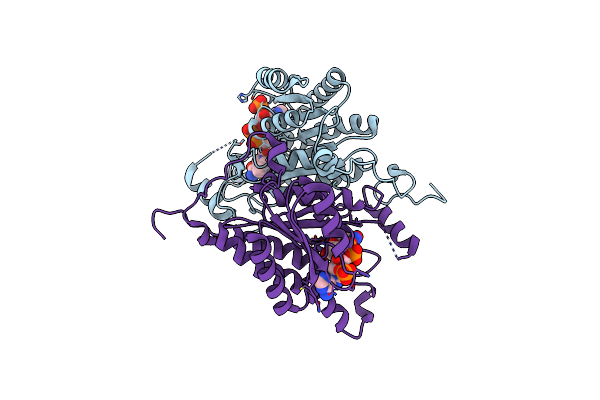 |
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: NDP |

