Search Count: 31
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-10 Classification: DNA BINDING PROTEIN |
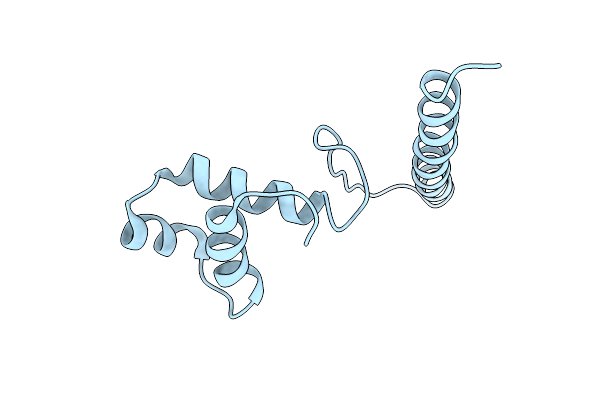 |
Crystal Structure Of The Hth Dna Binding Protein Ardk From R388 Plasmid. Apo Form.
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-07-13 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Hth Dna Binding Protein Ardk From R388 Plasmid Bound To A Direct-Repeat Dna Element
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-07-13 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Hth Dna Binding Protein Ardk From R388 Plasmid Bound To Ir3 Dna
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN |
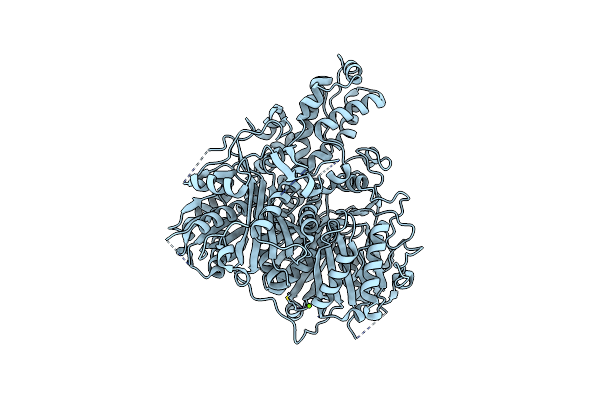 |
Organism: Moritella marina
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-08-26 Classification: ? Ligands: MG |
 |
Crystal Structure Of Antirestriction Ardc Protein From R388 Plasmid. Mn(Ii)-Bound Structure.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-15 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of Antirestriction Ardc Protein From R388 Plasmid. Metal-Free Structure.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-18 Classification: METAL BINDING PROTEIN Ligands: MPD |
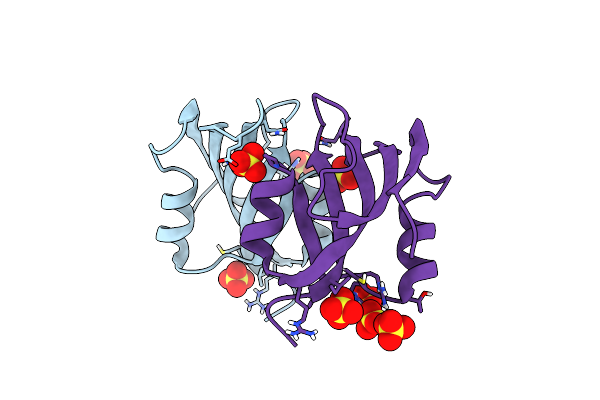 |
Crystal Structure Of The Master-Rep Protein Nuclease Domain From The Faba Bean Necrotic Yellows Virus
Organism: Faba bean necrotic yellows virus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2018-08-22 Classification: REPLICATION Ligands: SO4 |
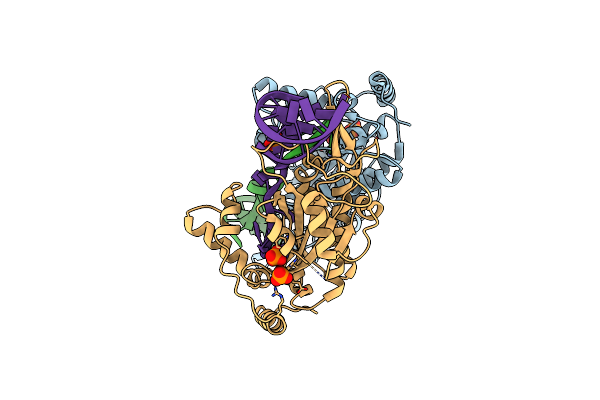 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-09-17 Classification: TRANSFERASE/DNA Ligands: PO4 |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2011-03-30 Classification: REPLICATION Ligands: PO4 |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-03-30 Classification: REPLICATION |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-03-30 Classification: REPLICATION Ligands: ANP, MG |
 |
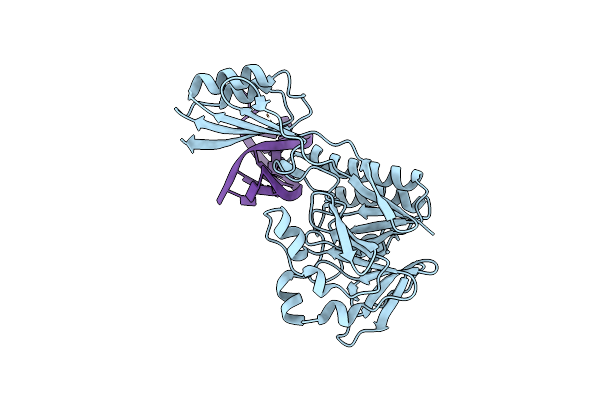
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2011-03-30 Classification: REPLICATION Ligands: ANP, MG |
 |
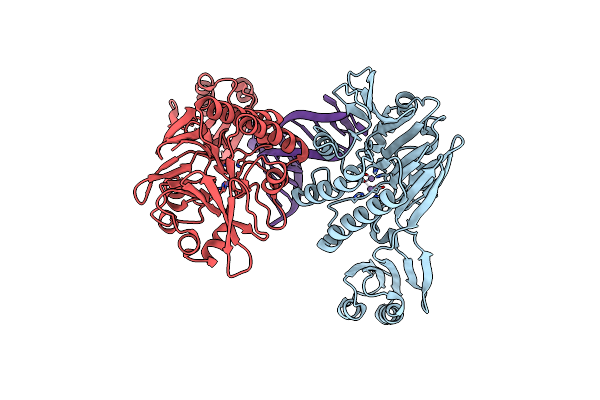
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-10-14 Classification: Hydrolase/DNA |
 |
Crystal Structure Of P. Furiosus Mre11-H85S Bound To A Branched Dna And Manganese
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-10-14 Classification: Hydrolase/DNA Ligands: MN |
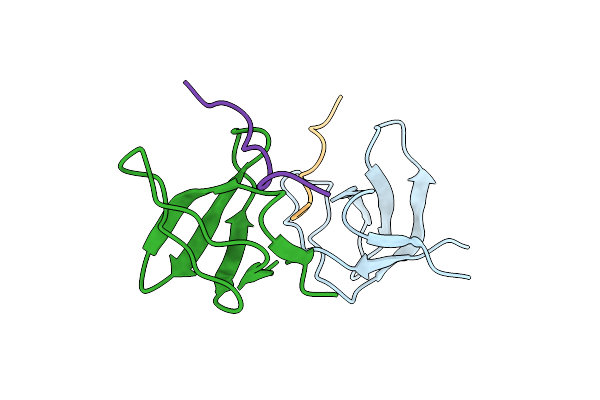 |
Atypical Polyproline Recognition By The Cms N-Terminal Sh3 Domain. Cms:Cd2 Heterodimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-11-06 Classification: PROTEIN BINDING |
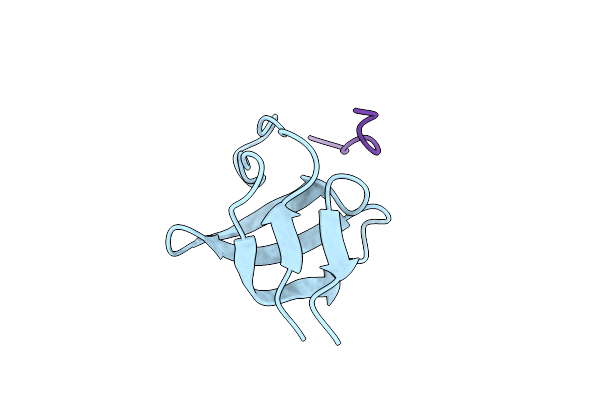 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-10-11 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2006-10-11 Classification: PROTEIN BINDING |
 |
Atypical Polyproline Recognition By The Cms N-Terminal Sh3 Domain. Cms:Cd2 Heterotrimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2006-10-11 Classification: PROTEIN BINDING |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2005-10-11 Classification: ENDOCYTOSIS/EXOCYTOSIS |

