Search Count: 44
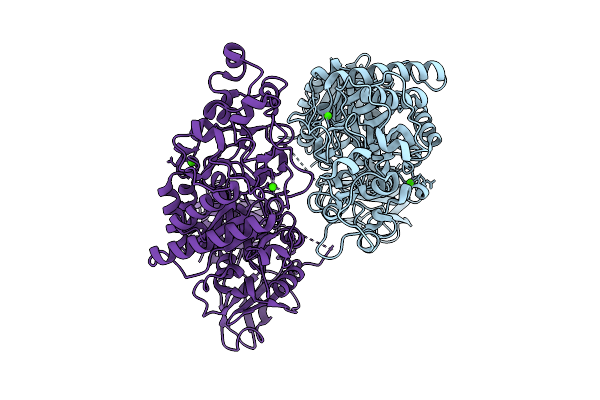 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA |
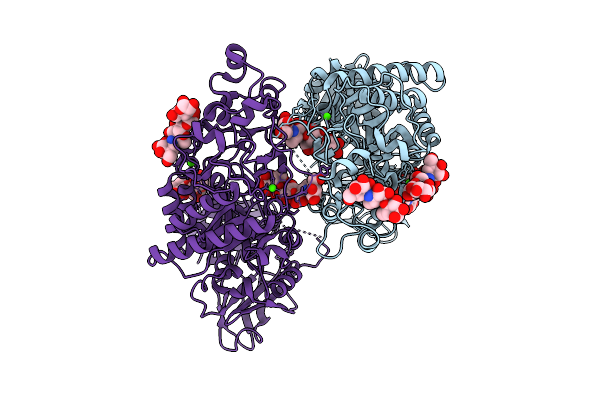 |
Klebsiella Pneumoniae Maltohexaose-Producing Alpha-Amylase In Complex With Acarbose
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA |
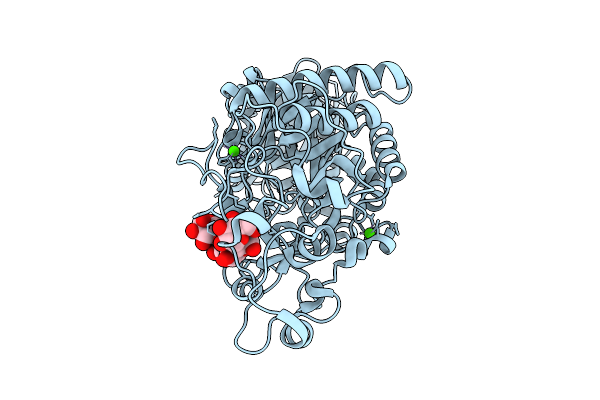 |
Domain N Deletion Mutant Of Klebsiella Pneumoniae Maltohexaose-Producing Alpha-Amylase In Complex With Maltohexaose
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA |
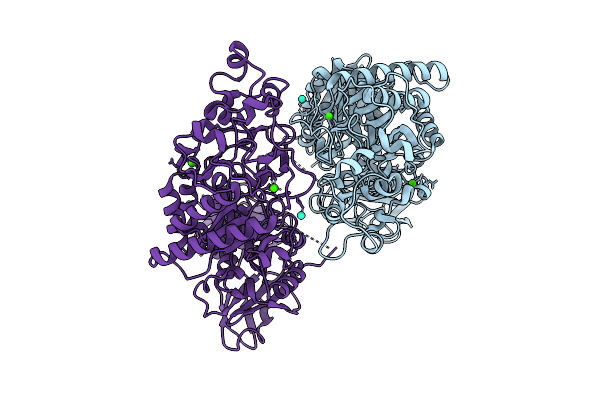 |
Klebsiella Pneumoniae Maltohexaose-Producing Alpha-Amylase, Terbium Derivative
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE Ligands: CA, TB |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: NAD, MG |
 |
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, NI, MG, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Alpha-1,6-Glucosyltransferase Complexed With Acarbose
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, NI, MG, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Alpha-1,6-Glucosyltransferase Complexed With Maltohexaose
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, BGC, GLC, NI, MG, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Alpha-1,6-Glucosyltransferase Complexed With Isomaltohexaose
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, BGC, GLC, NI, MG, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Alpha-1,6-Glucosyltransferase, Terbium Derivative
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-07-26 Classification: HYDROLASE, TRANSFERASE Ligands: CA, NI, MG, TB, SO4, MES, EDO |
 |
Crystal Structure Of Paenibacillus Sp. 598K Cycloisomaltooligosaccharide Glucanotransferase
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-04-26 Classification: TRANSFERASE Ligands: CA, NA, MLI, GOL |
 |
Crystal Structure Of Paenibacillus Sp. 598K Cycloisomaltooligosaccharide Glucanotransferase Complexed With Cycloisomaltoheptaose
Organism: Paenibacillus sp. 598k
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-04-26 Classification: TRANSFERASE Ligands: CA, NA, MLI |
 |
Crystal Structure Of Thermophilic Dextranase From Thermoanaerobacter Pseudethanolicus
Organism: Thermoanaerobacter pseudethanolicus atcc 33223
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: PO4, EDO, K, CXS |
 |
Crystal Structure Of Thermophilic Dextranase From Thermoanaerobacter Pseudethanolicus, D312G Mutant In Complex With Isomaltohexaose
Organism: Thermoanaerobacter pseudethanolicus atcc 33223
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: PO4, GOL |
 |
Crystal Structure Of Bacillus Circulans T-3040 Cycloisomaltooligosaccharide Glucanotransferase
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: CD, CA, NA, ACT, GOL |
 |
D308A Mutant Of Bacillus Circulans T-3040 Cycloisomaltooligosaccharide Glucanotransferase Complexed With Isomaltohexaose
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: CA, NA, SO4, MES |
 |
D308A Mutant Of Bacillus Circulans T-3040 Cycloisomaltooligosaccharide Glucanotransferase Complexed With Isomaltoheptaose
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: CA, NA, SO4, MES |
 |
D308A Mutant Of Bacillus Circulans T-3040 Cycloisomaltooligosaccharide Glucanotransferase Complexed With Isomaltooctaose
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: CA, NA, SO4, MES |
 |
D308A Mutant Of Bacillus Circulans T-3040 Cycloisomaltooligosaccharide Glucanotransferase Complexed With Cycloisomaltooctaose
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-02-05 Classification: TRANSFERASE Ligands: CA, NA, SO4, EDO, MES |

