Search Count: 17
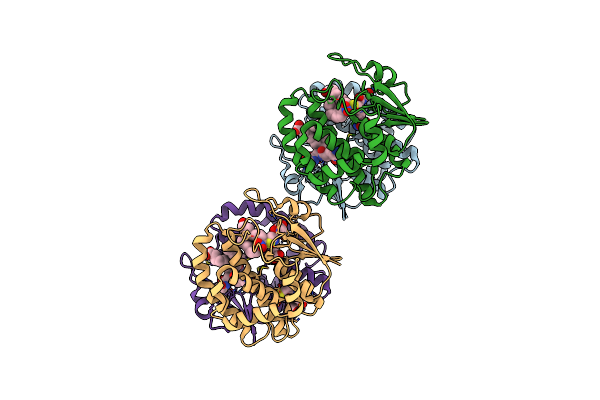 |
Human Glutathione Transferase M1-1 In Complex With The Adduct Between Glutathione And Nitrooleic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-03-12 Classification: TRANSFERASE/PRODUCT Ligands: A1AWW, GSH |
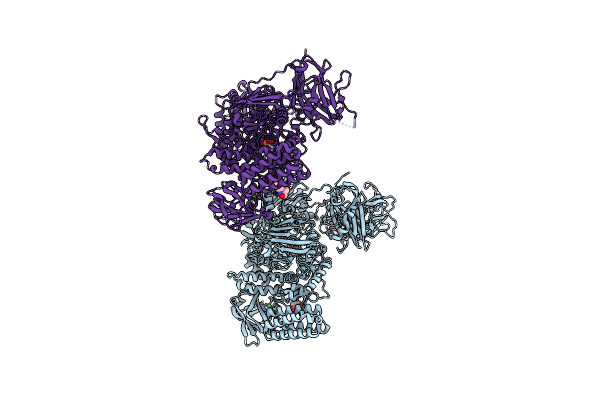 |
Multi-Domain Gh92 Alpha-1,2-Mannosidase From Neobacillus Novalis: Mannoimidazole Complex
Organism: Neobacillus novalis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: CA, MVL, B3P |
 |
Organism: Thermosynechococcus elongatus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: PHOTOSYNTHESIS Ligands: FE, MN, CL, PHO, CLA, BCR, LHG, BCT, LMG, PL9, HEM |
 |
Organism: Thermosynechococcus elongatus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: PHOTOSYNTHESIS Ligands: MN, CL, PHO, CLA, BCR, LHG, LMG, FE, PL9, HEM |
 |
Organism: Thermosynechococcus elongatus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2021-05-05 Classification: PHOTOSYNTHESIS Ligands: FE, MN, CL, PHO, CLA, BCR, LHG, LMG, PL9, HEM |
 |
Organism: Lactobacillus acidophilus ncfm
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2020-05-27 Classification: HYDROLASE Ligands: CA |
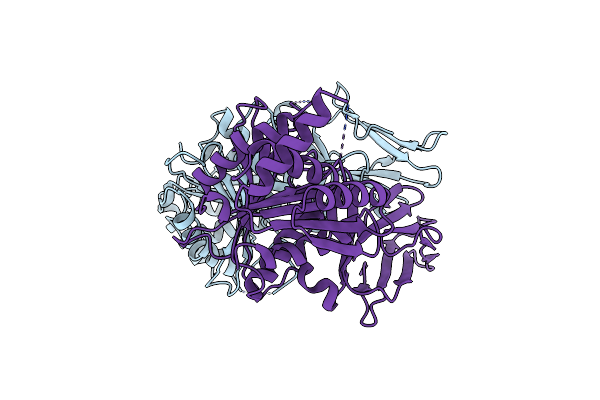 |
Crystal Structure Of The N-Terminal Carbohydrate Binding Module Family 48 And Ferulic Acid Esterase From The Multi-Enzyme Ce1-Gh62-Gh10
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-09-25 Classification: HYDROLASE |
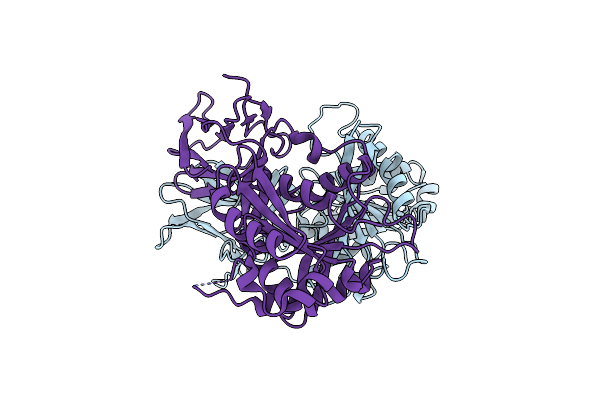 |
Crystal Structure Of The N-Terminal Carbohydrate Binding Module Family 48 And Ferulic Acid Esterase From The Multi-Enzyme Ce1-Gh62-Gh10
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2019-09-25 Classification: HYDROLASE |
 |
Structure Of The Barley Limit Dextrinase-Limit Dextrinase Inhibitor Complex
Organism: Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2015-04-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
Structure Of Glucan-1,6-Alpha-Glucosidase From Lactobacillus Acidophilus Ncfm
Organism: Lactobacillus acidophilus ncfm
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2012-08-29 Classification: HYDROLASE Ligands: MES, CA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2012-03-21 Classification: SIGNALING PROTEIN |
 |
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-27 Classification: OXIDOREDUCTASE Ligands: ZN, GOL, ACY |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-06-16 Classification: OXIDOREDUCTASE Ligands: CA, EDO |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-06-16 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2009-06-16 Classification: OXIDOREDUCTASE Ligands: EDO, UNX |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-06-16 Classification: OXIDOREDUCTASE Ligands: EDO, UNX |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-01-20 Classification: OXIDOREDUCTASE |

