Search Count: 87
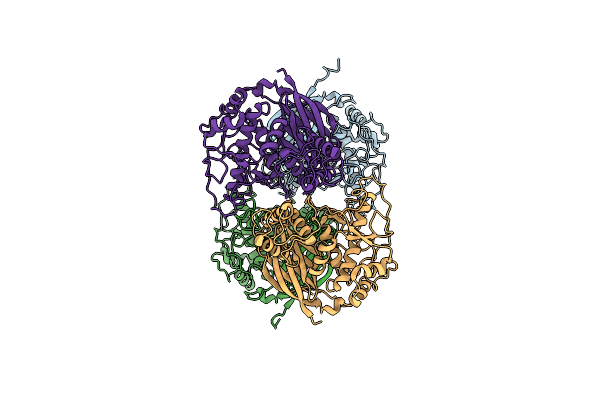 |
Crystal Structure Of The Spore Gernation Lytic Transglycosylase Slec From Clostridioides Difficile In Its Zymogenic Form (Prepro-Slec)
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-12 Classification: HYDROLASE |
 |
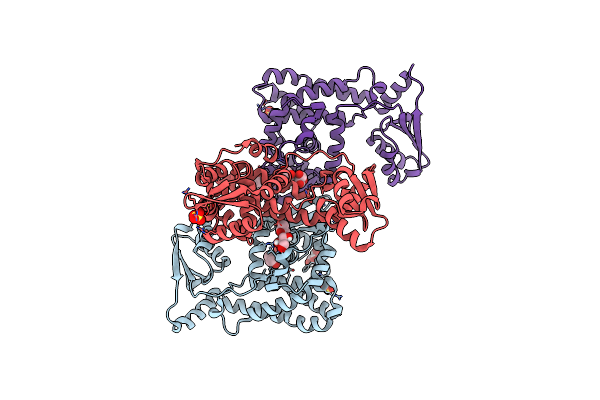
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: CYTOSOLIC PROTEIN Ligands: CA |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: CYTOSOLIC PROTEIN Ligands: CA |
 |
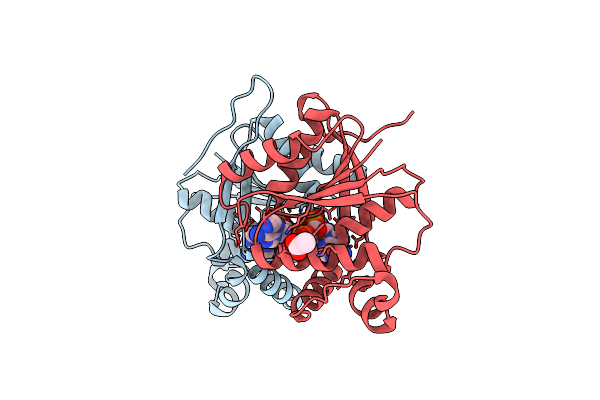
Organism: Escherichia coli w
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: GOL, SO4 |
 |
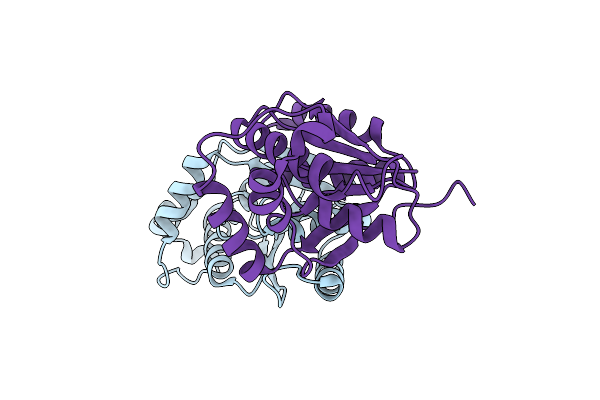
Crystal Structure Of The Ring Nuclease 0455 From Sulfolobus Islandicus (Sis0455) In Complex With Its Substrate
Organism: Sulfolobus islandicus rey15a, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-11-23 Classification: ANTIVIRAL PROTEIN Ligands: PO4, ACT |
 |
Crystal Structure Of The Ring Nuclease 0455 From Sulfolobus Islandicus (Sis0455) In Its Apo Form
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2022-11-23 Classification: ANTIVIRAL PROTEIN |
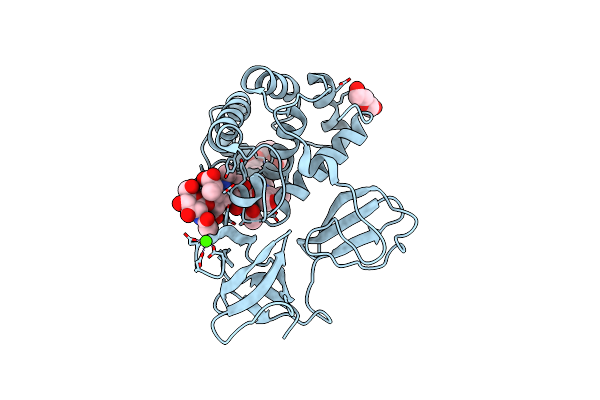 |
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
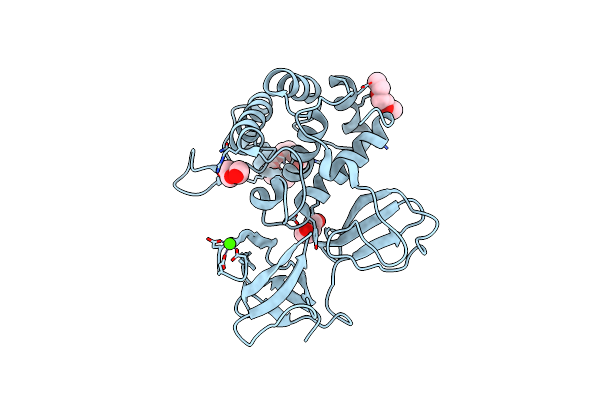 |
Crystal Structure Of Catalytic Domain In Open Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb (E585Q)From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, ACT, CA |
 |
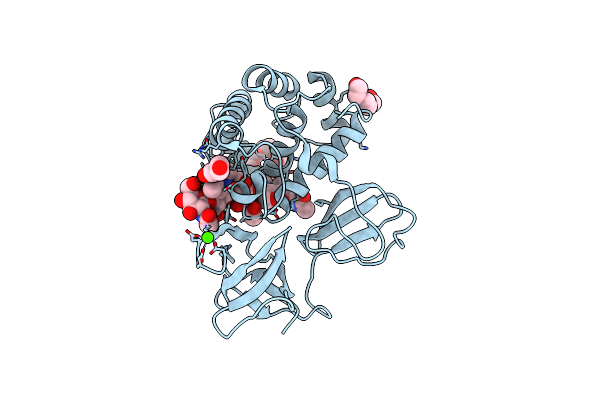
Crystal Structure Of Catalytic Domain Of Lytb From Streptococcus Pneumoniae In Complex With Nag-Nag-Nag-Nag Tetrasaccharide
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
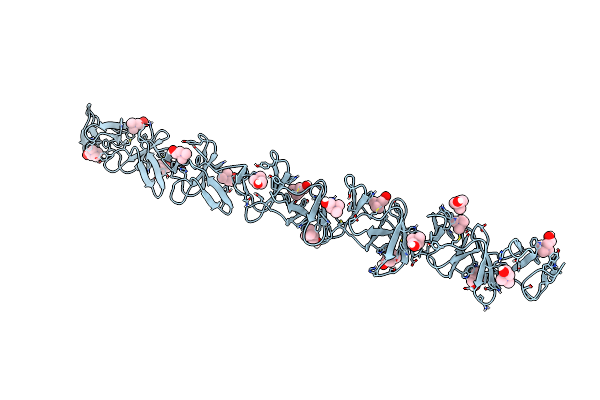
Crystal Structure Of Catalytic Domain Of Lytb (E585Q) From Streptococcus Pneumoniae In Complex With Nag-Nam-Nag-Nam-Nag Peptidolycan Analogue
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PGE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT |
 |
Crystal Structure Of Catalytic Domain In Closed Conformation Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: 1PE, PEG, CA |
 |
Crystal Structure Of Choline-Binding Module (R1-R9) Of Lytb From Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae (strain atcc baa-255 / r6)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: CHT, ZN, PGE |
 |
Organism: Aequorea victoria, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN Ligands: K |
 |
Organism: Thermus thermophilus hb8, Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-12-22 Classification: FLUORESCENT PROTEIN Ligands: CA, 2OP |
 |
Crystal Structure Of The Ring Nuclease 0811 Mutant-S12G/K169G From Sulfolobus Islandicus (Sis0811)
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-12-01 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of The Ring Nuclease 0811 From Sulfolobus Islandicus (Sis0811) In Its Apo Form
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2021-11-24 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of The Ring Nuclease 0811 From Sulfolobus Islandicus (Sis0811) In Complex With Its Post-Catalytic Reaction Product
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-11-24 Classification: ANTIVIRAL PROTEIN Ligands: LQJ |
 |
Crystal Structure Of The Ring Nuclease 0811 Mutant-S12A From Sulfolobus Islandicus (Sis0811)
Organism: Sulfolobus islandicus rey15a
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2021-11-24 Classification: ANTIVIRAL PROTEIN |

