Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
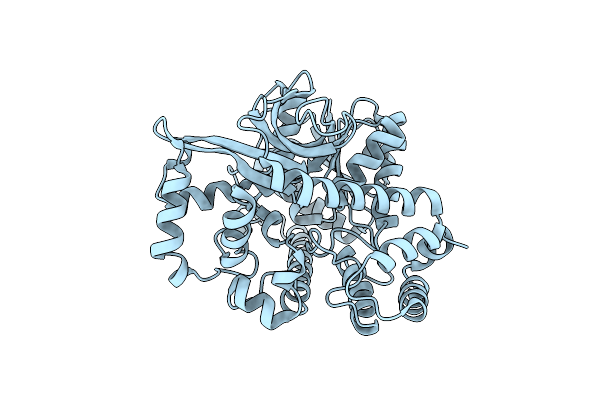 |
Crystal Structure Of Apo Form Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-09-01 Classification: HYDROLASE |
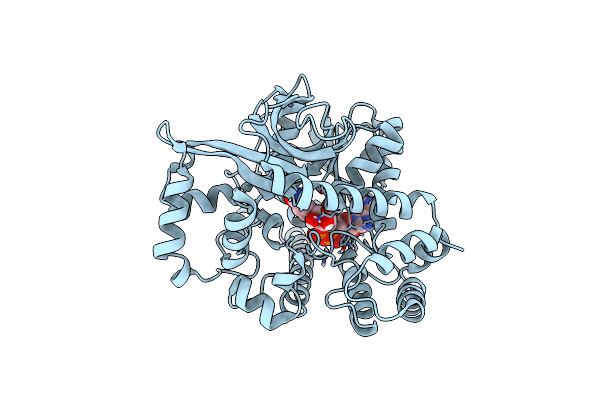 |
Crystal Structure Of Nadh Bound Holo Form Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima At 1.97 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI |
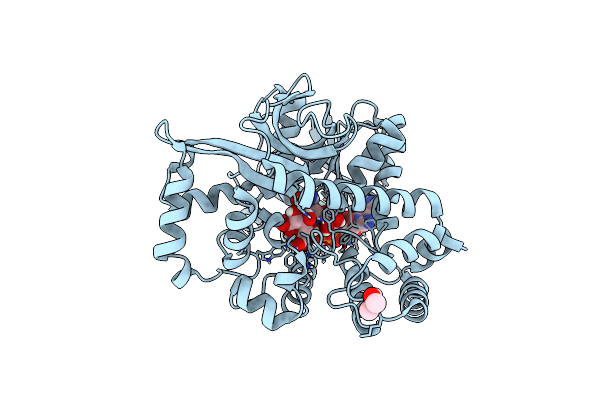 |
Crystal Structure Of Thermotoga Maritima Alpha-Glucuronidase (Tm0752) In Complex With Nadh And D-Glucuronic Acid
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI, BDP, IPA |
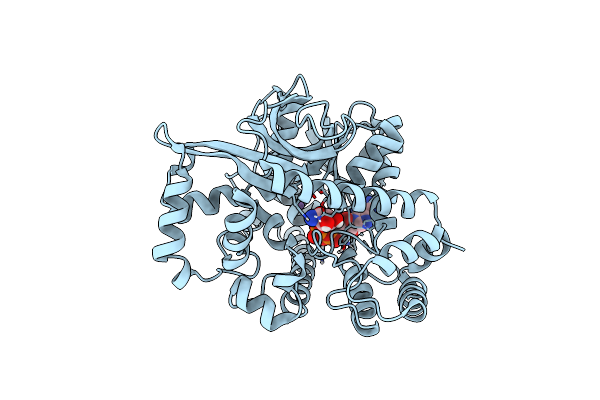 |
Structure Of Deletion Mutant Of Alpha-Glucuronidase (Tm0752) From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: MN, NAI |
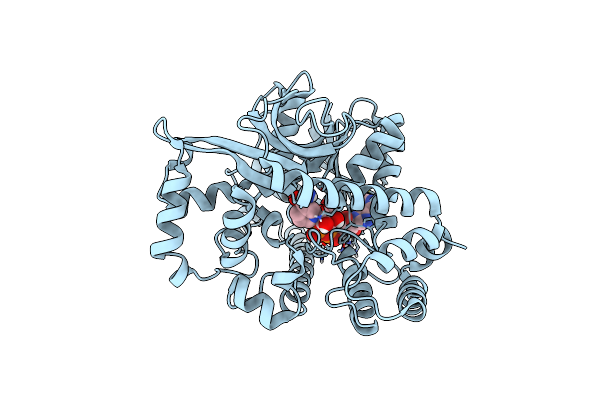 |
Structure Of Nadh Complex Of Thermotoga Maritima Alpha-Glucuronidase At 2.15 Angstrom Resolution
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-31 Classification: HYDROLASE Ligands: NAI |
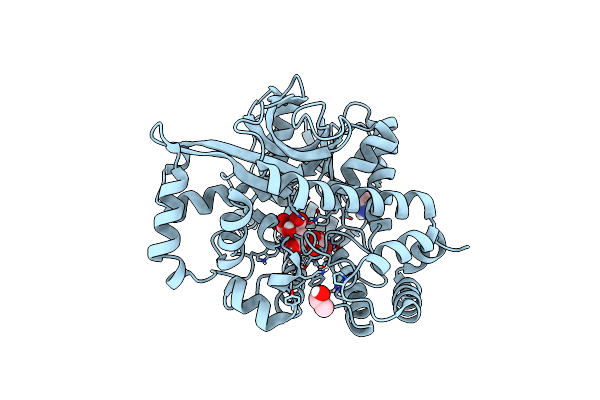 |
Crystal Structure Of Citrate Complex Of Alpha-Glucuronidase (Tm0752)From Thermotoga Maritima
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: CO, IMD, IPA, CIT |
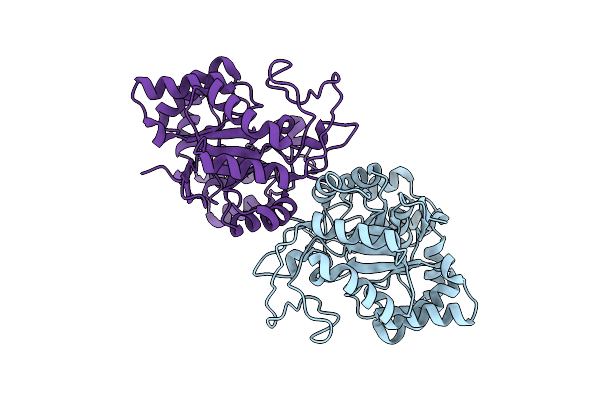 |
Crystal Structure Of Apo Form Of Xylose Reductase From Debaryomyces Nepalensis
Organism: Debaryomyces nepalensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE |
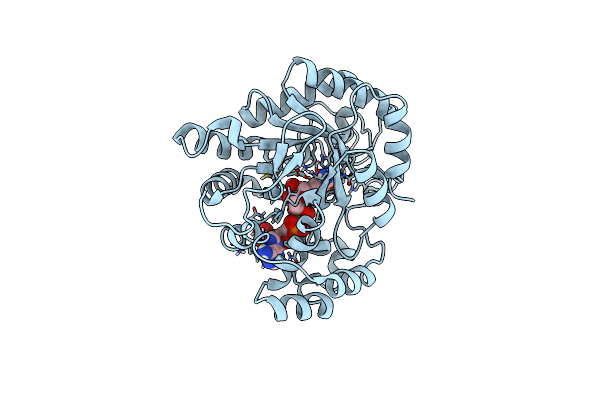 |
Crystal Structure Of Xylose Reductase From Debaryomyces Nepalensis In Complex With Nadp-Dtt Adduct
Organism: Debaryomyces nepalensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: NDP, DTT |