Search Count: 32
 |
Crystal Structure Determination Of Dye-Decolorizing Peroxidase (Dyp) From Deinoccoccus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN Ligands: HEM, GOL, MG |
 |
Crystal Structure Determination Of Dye-Decolorizing Peroxidase (Dyp) Mutant M190G From Deinoccoccus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN Ligands: HEM, OH, CL, CA |
 |
Ferric-Siderophore Reduction In Shewanella Biscestrii: Structural And Functional Characterization Of Sbisip Reveals Unforeseen Specificity
Organism: Shewanella bicestrii
Method: X-RAY DIFFRACTION Release Date: 2024-01-17 Classification: METAL BINDING PROTEIN Ligands: FAD |
 |
Structure Of The C-Terminally Truncated Nad+-Dependent Dna Ligase From The Poly-Extremophile Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2023-09-27 Classification: DNA BINDING PROTEIN Ligands: MN, ZN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: FES |
 |
Crystal Structure Of Deinococcus Radiodurans Endonuclease Iii-1 Y100L Variant
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-08-03 Classification: LYASE Ligands: SF4, MG |
 |
Crystal Structure Of Deinococcus Radiodurans Endonuclease Iii-1 R61Q Variant
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-08-03 Classification: LYASE Ligands: MG, SF4 |
 |
Crystal Structure Of Deinococcus Radiodurans Endonuclease Iii-3 Double Mutant
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-08-03 Classification: LYASE Ligands: SF4, CL |
 |
Organism: Shewanella algae
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2019-10-16 Classification: ELECTRON TRANSPORT Ligands: HEC, PO3 |
 |
Structure And Reactivity Of A Siderophore-Interacting Protein From The Marine Bacterium Shewanella Reveals Unanticipated Functional Versatility.
Organism: Shewanella frigidimarina (strain ncimb 400)
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2018-11-21 Classification: METAL TRANSPORT Ligands: MG, DMS, FMT, NA, GOL, FAD |
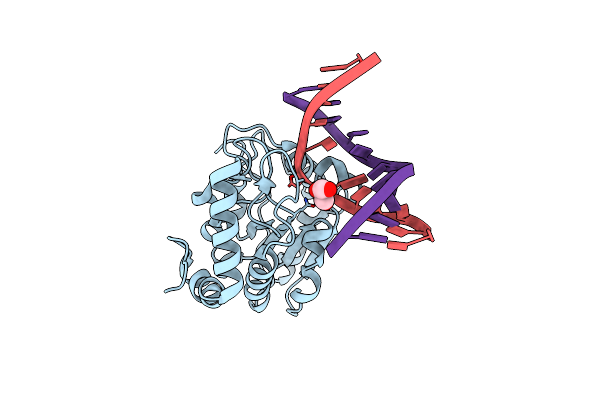 |
Crystal Structure Determination Of Uracil-Dna N-Glycosylase (Ung) From Deinococcus Radiodurans In Complex With Dna - New Insights Into The Role Of The Leucine-Loop For Damage Recognition And Repair
Organism: Deinococcus radiodurans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-08-12 Classification: HYDROLASE/DNA Ligands: CL, GOL |
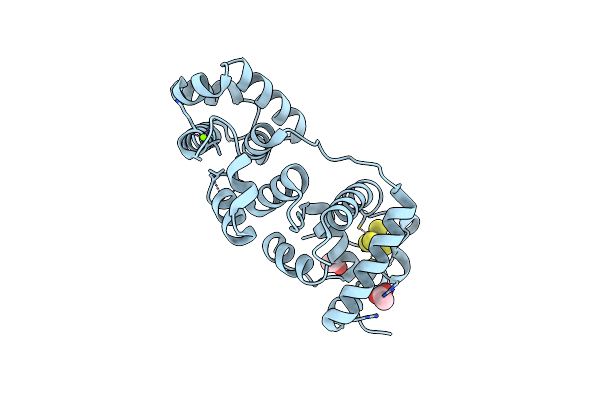 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-06-10 Classification: LYASE Ligands: SF4, MG, ACT |
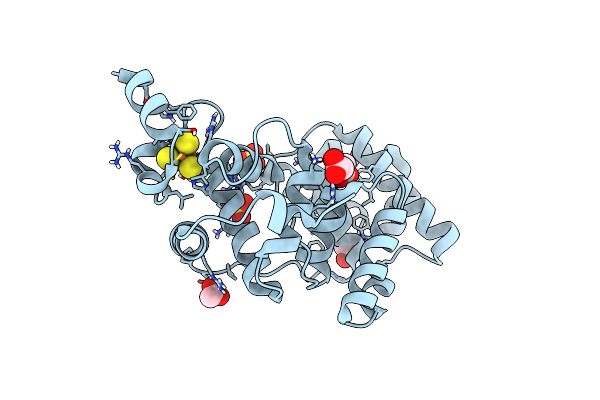 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2015-06-10 Classification: LYASE Ligands: SF4, CO, SO4, ACT |
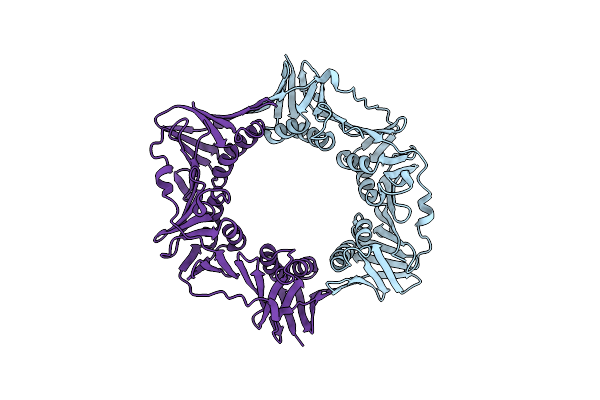 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-29 Classification: TRANSFERASE |
 |
Structure And Function Analysis Of Mutt From The Psychrofile Fish Pathogen Aliivibrio Salmonicida And The Mesophile Vibrio Cholerae
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2015-02-04 Classification: HYDROLASE |
 |
Crystal Structure Of Uracil-Dna Glycosylase From Cod (Gadus Morhua) In Complex With The Proteinaceous Inhibitor Ugi
Organism: Gadus morhua, Bacillus phage pbs2
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-08-13 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: IMD |
 |
Structure Of An Unusual 3-Methyladenine Dna Glycosylase Ii (Alka) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-20 Classification: HYDROLASE Ligands: CL, GOL, MES |
 |
Structure Of An Unusual 3-Methyladenine Dna Glycosylase Ii (Alka) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2011-04-20 Classification: HYDROLASE Ligands: CL, GOL, NI |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-06-03 Classification: HYDROLASE Ligands: CL |

