Search Count: 63
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
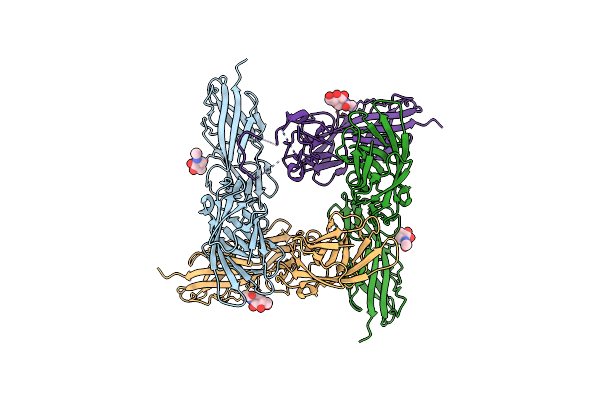 |
Structure-Based Engineering Of A Highly Immunogenic, Conformationally Stabilized Fimh Antigen For A Urinary Tract Infection Vaccine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-05 Classification: CELL ADHESION Ligands: NAG |
 |
Cryo-Em Structure Of E. Coli Fimh Lectin Domain Bound To Fabs 440-2 And 454-3
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: CELL ADHESION,SUGAR BINDING PROTEIN |
 |
Cryo-Em Structure Of E. Coli Fimh Lectin Domain Bound To Fabs 329-2 And 454-3
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CELL ADHESION/IMMUNE SYSTEM |
 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-06 Classification: STRUCTURAL PROTEIN Ligands: GOL, ATP, CA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2021-09-01 Classification: NUCLEAR PROTEIN Ligands: SAH, A2M |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-09-01 Classification: NUCLEAR PROTEIN Ligands: SAM |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2021-09-01 Classification: NUCLEAR PROTEIN Ligands: SFG, GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2021-09-01 Classification: NUCLEAR PROTEIN Ligands: SAH, GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-09-01 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-04-21 Classification: TRANSFERASE/Inhibitor Ligands: XRM, CL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2021-04-21 Classification: TRANSFERASE/Inhibitor Ligands: YFG, CL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2021-03-24 Classification: TRANSFERASE/Inhibitor Ligands: XBA, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-03-24 Classification: TRANSFERASE/TRANSFERASE Inhibitor Ligands: TZ7, EDO, PEG, GOL, SO4, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-03-24 Classification: TRANSFERASE/TRANSFERASE Inhibitor Ligands: FDR, EDO, TYG, GOL |
 |
Human Poly(Adp-Ribose) Polymerase 12, Catalytic Fragment With Four Point Mutations In Complex With Rbn-2397
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-12-16 Classification: TRANSFERASE Ligands: QO4, CL |

