Search Count: 16
 |
Solution Nmr Structure Of The Full Length Latent Form Mine Protein From Neisseria Gonorrheae
Organism: Neisseria gonorrhoeae
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: CELL CYCLE |
 |
Solution Nmr Structure Of The Partially Activated Mts Deleted Form Mine Protein (Delta10-Ngmine) From Neisseria Gonorrheae
Organism: Neisseria gonorrhoeae
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: CELL CYCLE |
 |
Solution Nmr Structure Of The Delta30-Ngmine Protein From Neisseria Gonorrheae
Organism: Neisseria gonorrhoeae
Method: SOLID-STATE NMR Release Date: 2020-07-08 Classification: CELL CYCLE |
 |
Solution Nmr Structure Of The I24N-Delta10-Ngmine Protein From Neisseria Gonorrheae
Organism: Neisseria gonorrhoeae
Method: SOLUTION NMR Release Date: 2020-07-08 Classification: CELL CYCLE |
 |
Mub Is An Aaaplus Atpase That Forms Helical Filaments To Control Target Selection For Dna Transposition
Organism: Enterobacteria phage mu
Method: ELECTRON MICROSCOPY Resolution:18.00 Å Release Date: 2013-07-03 Classification: HYDROLASE Ligands: ADP |
 |
Mub Is An Aaaplus Atpase That Forms Helical Filaments To Control Target Selection For Dna Transposition
Organism: Enterobacteria phage mu
Method: ELECTRON MICROSCOPY Resolution:17.00 Å Release Date: 2013-07-03 Classification: TRANSCRIPTION Ligands: ADP |
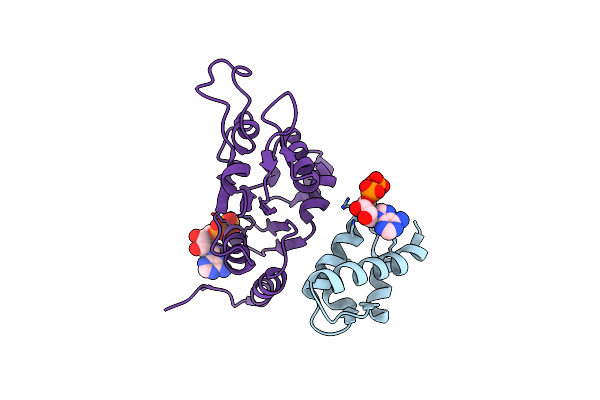 |
Mub Is An Aaaplus Atpase That Forms Helical Filaments To Control Target Selection For Dna Transposition
Organism: Enterobacteria phage mu
Method: ELECTRON MICROSCOPY Resolution:16.00 Å Release Date: 2013-07-03 Classification: TRANSCRIPTION Ligands: ADP |
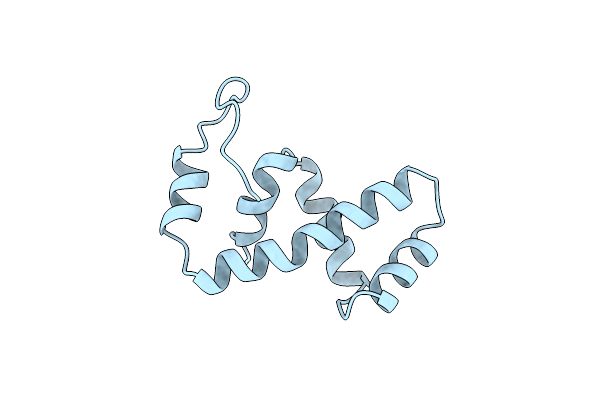 |
Solution Nmr Structure Of The Ibeta Subdomain Of The Mu End Dna Binding Domain Of Phage Mu Transposase, Regularized Mean Structure
Organism: Enterobacteria phage mu
Method: SOLUTION NMR Release Date: 1998-01-14 Classification: DNA BINDING PROTEIN |
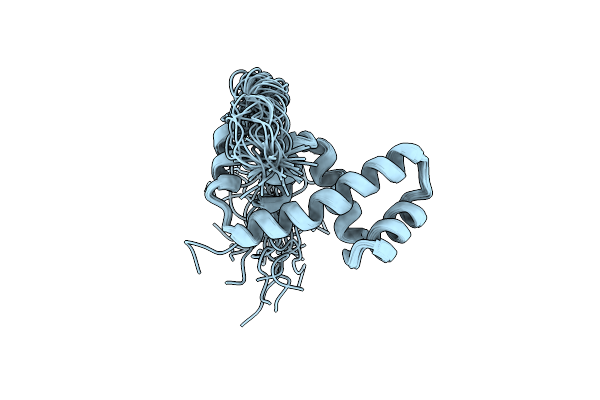 |
Solution Nmr Structure Of The Ibeta Subdomain Of The Mu End Dna Binding Domain Of Phage Mu Transposase, 29 Structures
Organism: Enterobacteria phage mu
Method: SOLUTION NMR Release Date: 1998-01-14 Classification: DNA BINDING PROTEIN |
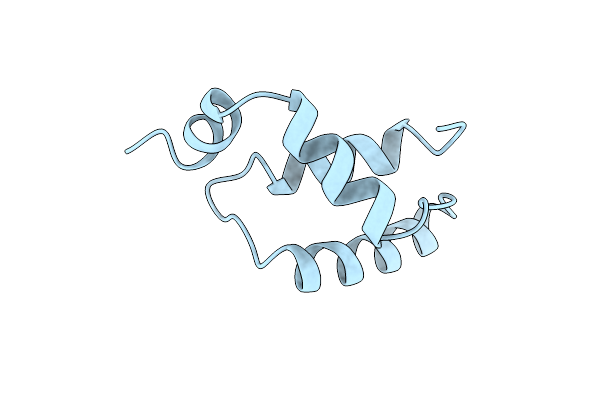 |
Solution Nmr Structure Of The Igamma Subdomain Of The Mu End Dna Binding Domain Of Mu Phage Transposase, Minimized Average Structure
Organism: Enterobacteria phage mu
Method: SOLUTION NMR Release Date: 1997-12-03 Classification: DNA BINDING PROTEIN |
 |
Solution Nmr Structure Of The Igamma Subdomain Of The Mu End Dna Binding Domain Of Mu Phage Transposase, 30 Structures
Organism: Enterobacteria phage mu
Method: SOLUTION NMR Release Date: 1997-12-03 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1997-02-12 Classification: TRANSCRIPTION/DNA Ligands: CD |
 |
Bacteriophage Mu Transposase Core Domain With 2 Monomers Per Asymmetric Unit
Organism: Enterobacteria phage mu
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1995-10-15 Classification: TRANSPOSASE |
 |
Organism: Enterobacteria phage mu
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1995-10-15 Classification: TRANSPOSASE |
 |
A Novel Class Of Winged Helix-Turn-Helix Protein: The Dna-Binding Domain Of Mu Transposase
Organism: Enterobacteria phage mu
Method: SOLUTION NMR Release Date: 1995-02-14 Classification: DNA BINDING PROTEIN |
 |
A Novel Class Of Winged Helix-Turn-Helix Protein: The Dna-Binding Domain Of Mu Transposase
Organism: Enterobacteria phage mu
Method: SOLUTION NMR Release Date: 1995-02-14 Classification: DNA BINDING PROTEIN |

