Search Count: 179
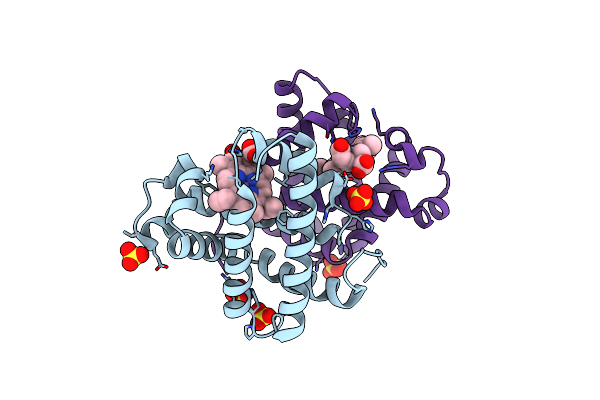 |
Crystal Structure Of Sperm Whale Myoglobin Reconstituted With Manganese Porphycene
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-08-21 Classification: OXYGEN STORAGE Ligands: HNN, SO4 |
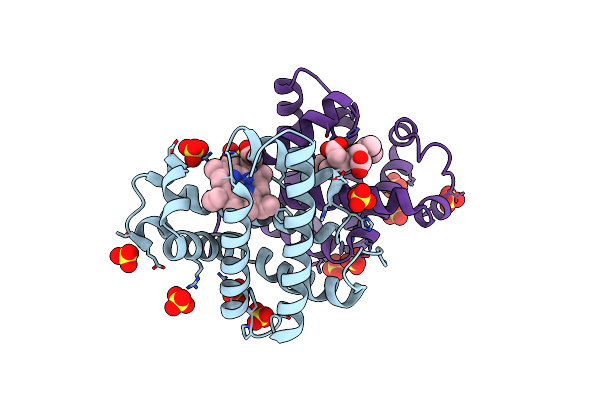 |
Crystal Structure Of Sperm Whale Myoglobin (F43A/H64I Mutant) Reconstituted With Manganese Porphycene
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-08-21 Classification: OXYGEN STORAGE Ligands: HNN, SO4 |
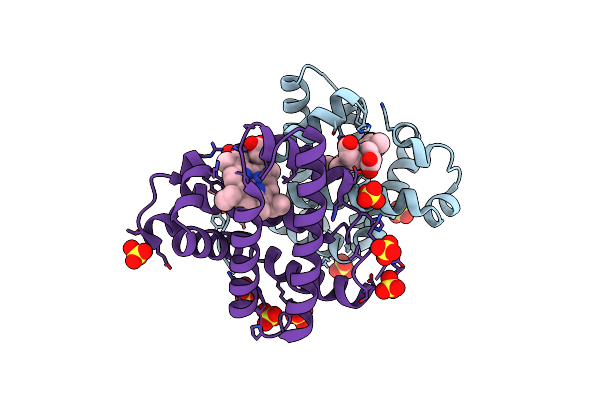 |
Crystal Structure Of Sperm Whale Myoglobin (F46L/H64A Mutant) Reconstituted With Manganese Porphycene
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-08-21 Classification: OXYGEN STORAGE Ligands: HNN, SO4, CL |
 |
Cryo-Em Structure Of Groel Bound To Unfolded Substrate (Ugt1A) At 2.8 Ang. Resolution (Consensus Refinement)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Double Occupied Ring (Dor) Of Groel-Ugt1A Complex At 2.7 Ang. Resolution
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Single Empty Ring 2 (Ser2) Of Groel-Ugt1A Complex At 3.2 Ang. Resolution
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 4 (Or4) Of Groel Complexed With Polyalanine Model Of Ugt1A From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Empty Ring Subunit 1 (Er1) From Single Empty Ring Of Groel-Ugt1A Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Empty Ring Subunit 2 (Er2) From Groel-Ugt1A Single Empty Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 1 (Or1) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 2 (Or2) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Occupied Ring Subunit 3 (Or3) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
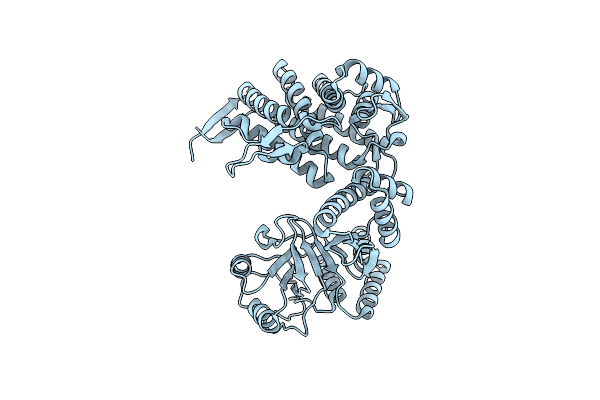 |
Cryo-Em Structure Of Occupied Ring Subunit 4 (Or4) Of Groel From Groel-Ugt1A Double Occupied Ring Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: CHAPERONE |
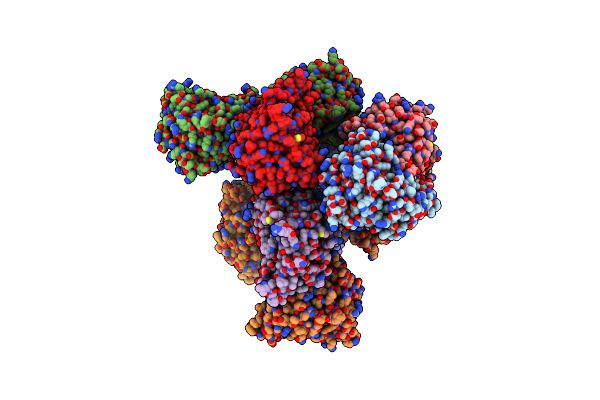 |
Crystal Structure Of The Aminopropyltransferase, Spee From Hyperthermophilic Crenarchaeon, Pyrobaculum Calidifontis In Complex With 5'-Methylthioadenosine (Mta) Alone Or Together With Spermidine Or Thermospermine
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MTA, SPD, TER |
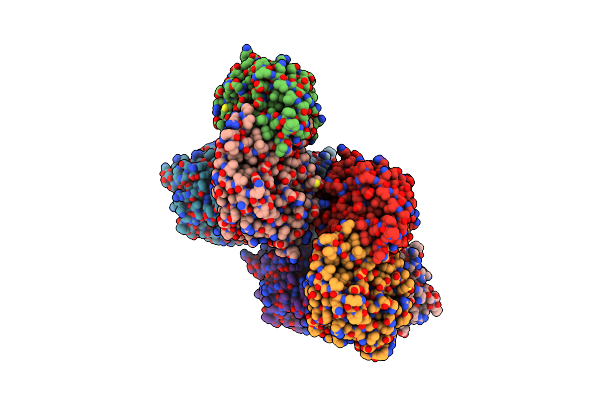 |
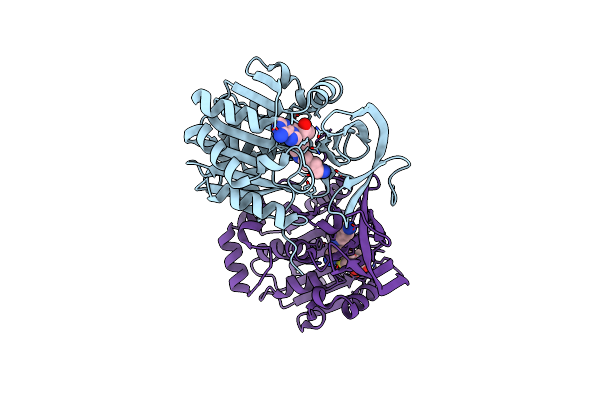
Crystal Structure Of The Aminopropyltransferase, Spee From Hyperthermophilic Crenarchaeon, Pyrobaculum Calidifontis In Complex With 5'-Methylthioadenosine (Mta) And Spermine
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MTA, SPM, EPE |
 |
Crystal Structure Of The Aminopropyltransferase, Spee From Hyperthermophilic Crenarchaeon, Pyrobaculum Calidifontis In Complex With 5'-Methylthioadenosine (Mta) And Spermidine
Organism: Pyrobaculum calidifontis jcm 11548
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MTA, SPD |
 |
Crystal Structure Of The Aminopropyltransferase, Spee From Hyperthermophilic Crenarchaeon, Pyrobaculum Calidifontis In Complex With 5'-Methylthioadenosine (Mta) & Aminopropylagmatine
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-15 Classification: TRANSFERASE Ligands: MTA, AG3, EPE |
 |
Serial Femtosecond Crystallography (Sfx) Of Ground State Bacteriorhodopsin Crystallized From Bicelles In Complex With Had16 Determined Using 7-Kev X-Ray Free Electron Laser (Xfel) At Sacla
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, 7YH, IOD, CPS, OCT, HP6, D12, R16, D10, DD9 |
 |
Crystal Structure Of The Branched-Chain Polyamine Synthase From Thermococcus Kodakarensis (Tk-Bpsa) In Complex With N4-Bis(Aminopropyl)Spermidine And 5'-Methylthioadenosine
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: MTA, B5L, FE |
 |
Crystal Structure Of The Branched-Chain Polyamine Synthase From Thermus Thermophilus (Tth-Bpsa) In Complex With N4-Aminopropylspermidine And 5'-Methylthioadenosine
Organism: Thermus thermophilus (strain hb27 / atcc baa-163 / dsm 7039)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2019-06-26 Classification: TRANSFERASE Ligands: FE, MTA, N4P, PGE, PEG |

