Search Count: 26
 |
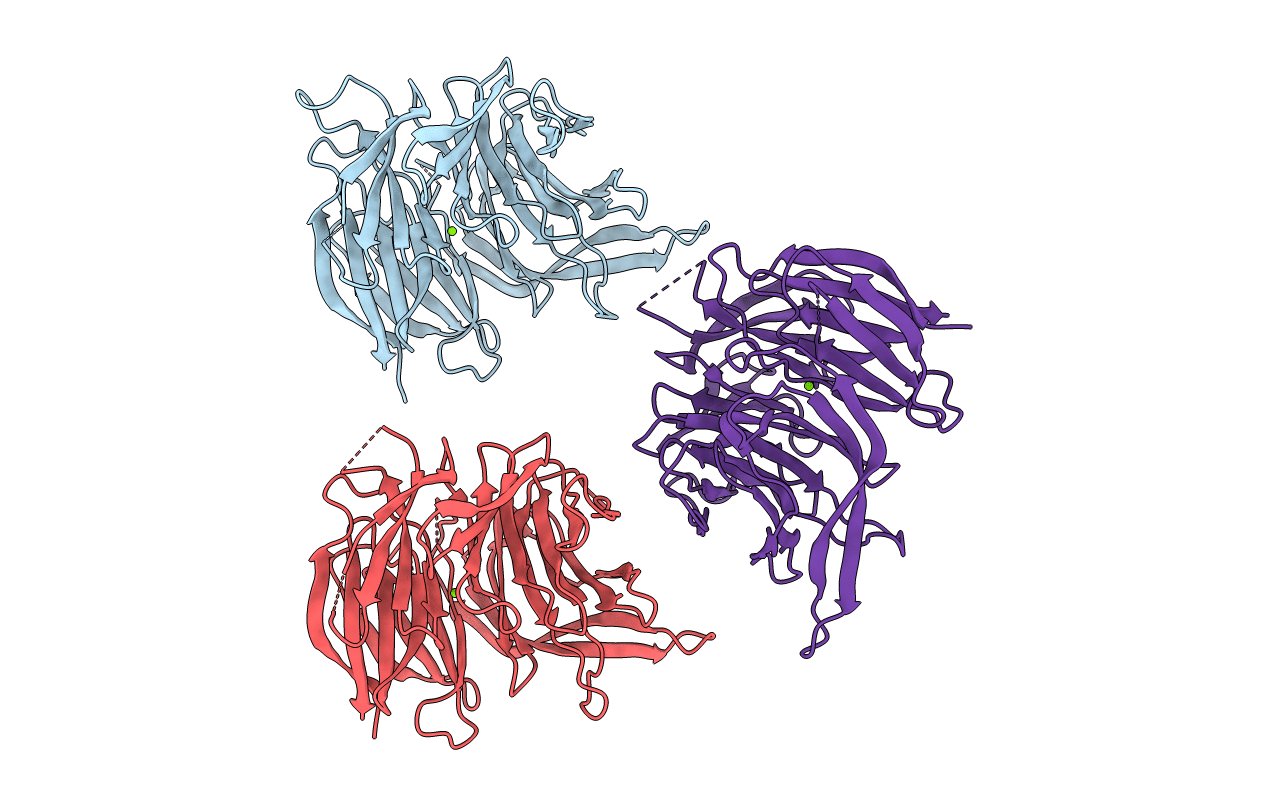
Organism: Bdellovibrio bacteriovorus (strain atcc 15356 / dsm 50701 / ncib 9529 / hd100)
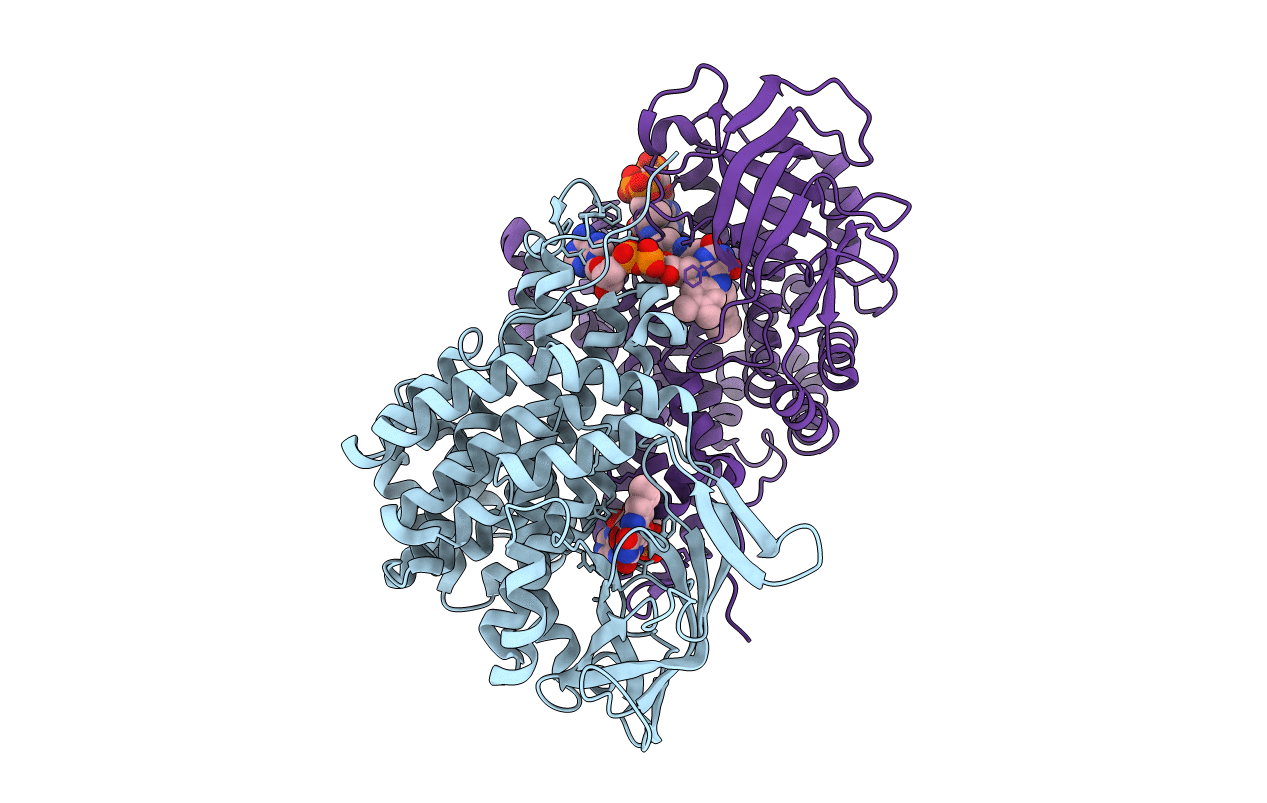
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2019-09-11 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2019-09-11 Classification: OXIDOREDUCTASE Ligands: FAD, MFK |
 |
Cryo-Em Structure Of The Anti-Feeding Prophage (Afp) Baseplate In Extended State, 3-Fold Symmetrised
Organism: Serratia entomophila
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The Anti-Feeding Prophage (Afp) Helical Sheath-Tube Complex In Extended State
Organism: Serratia entomophila
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRUS LIKE PARTICLE |
 |
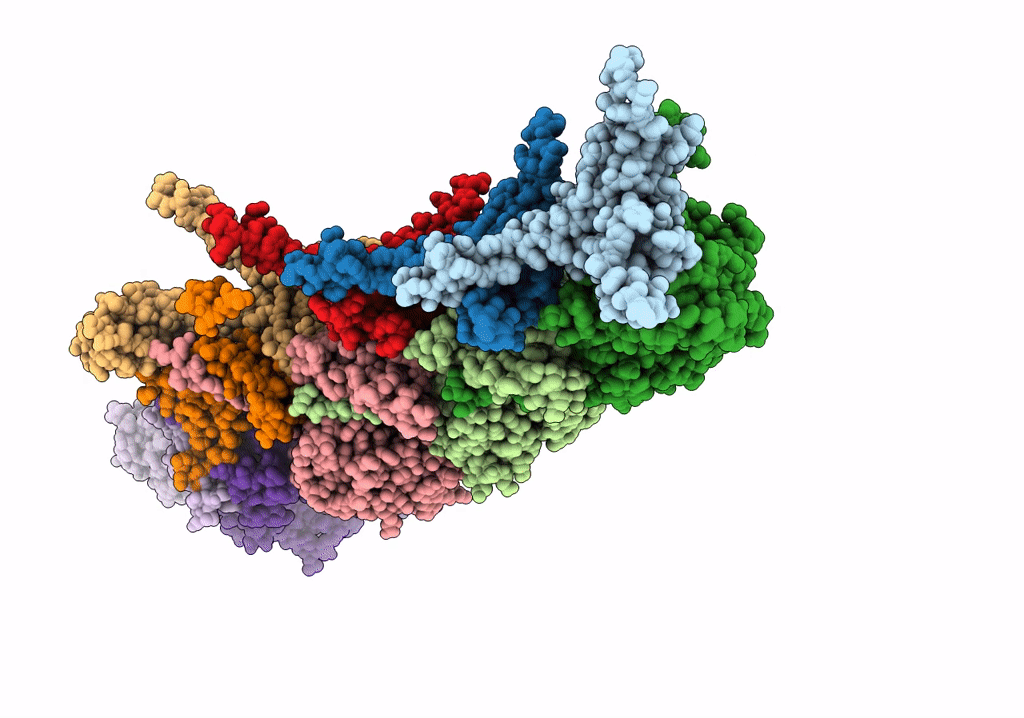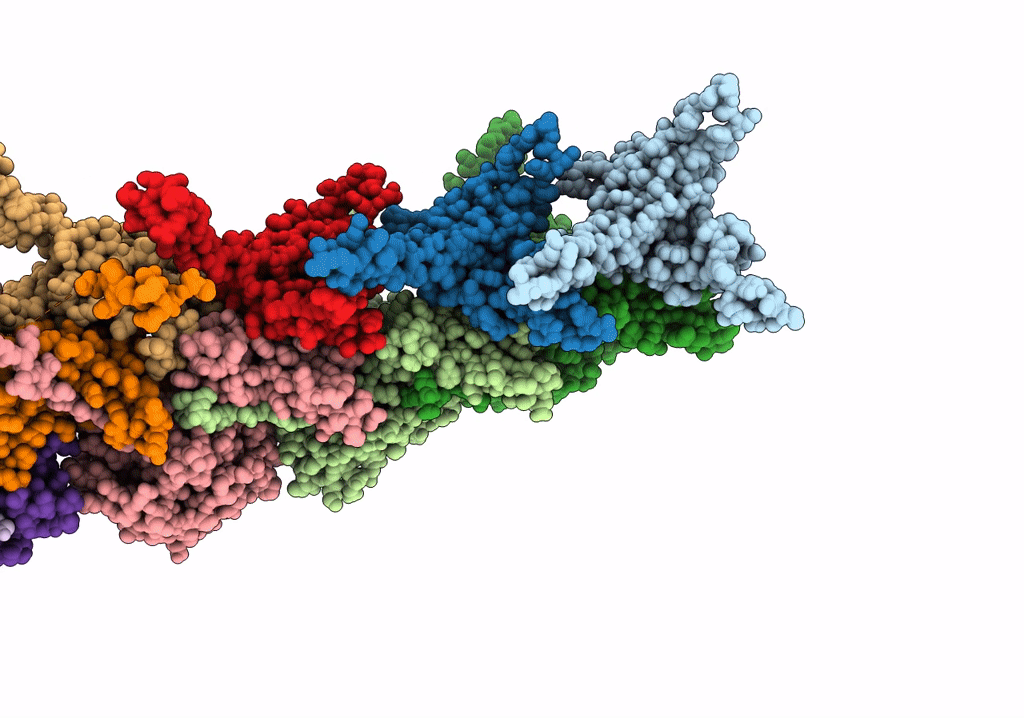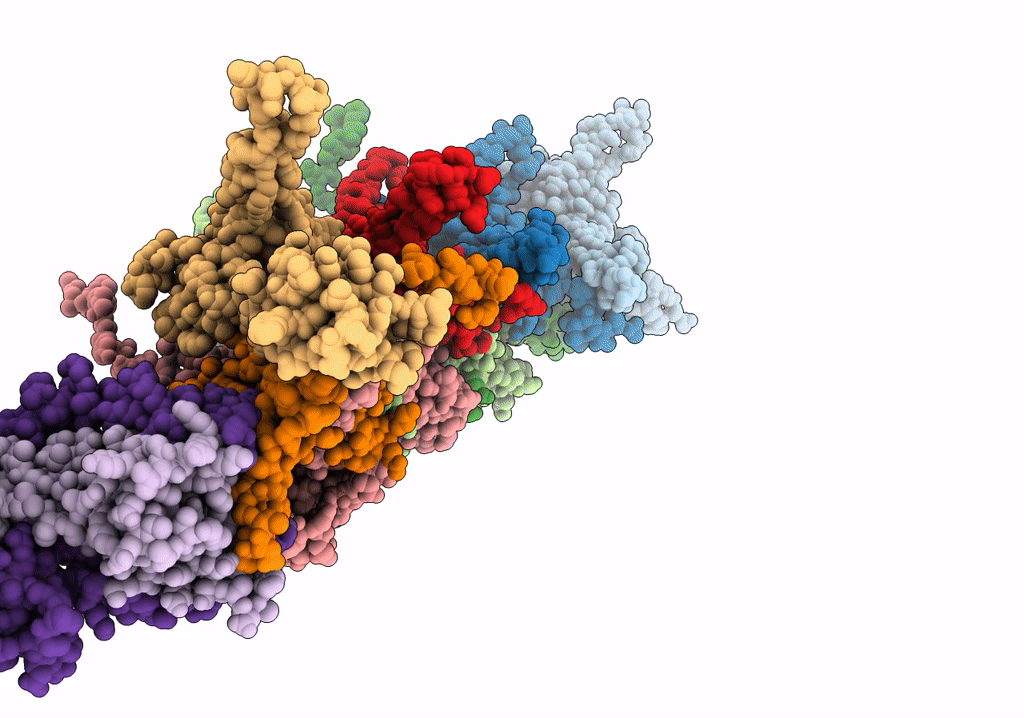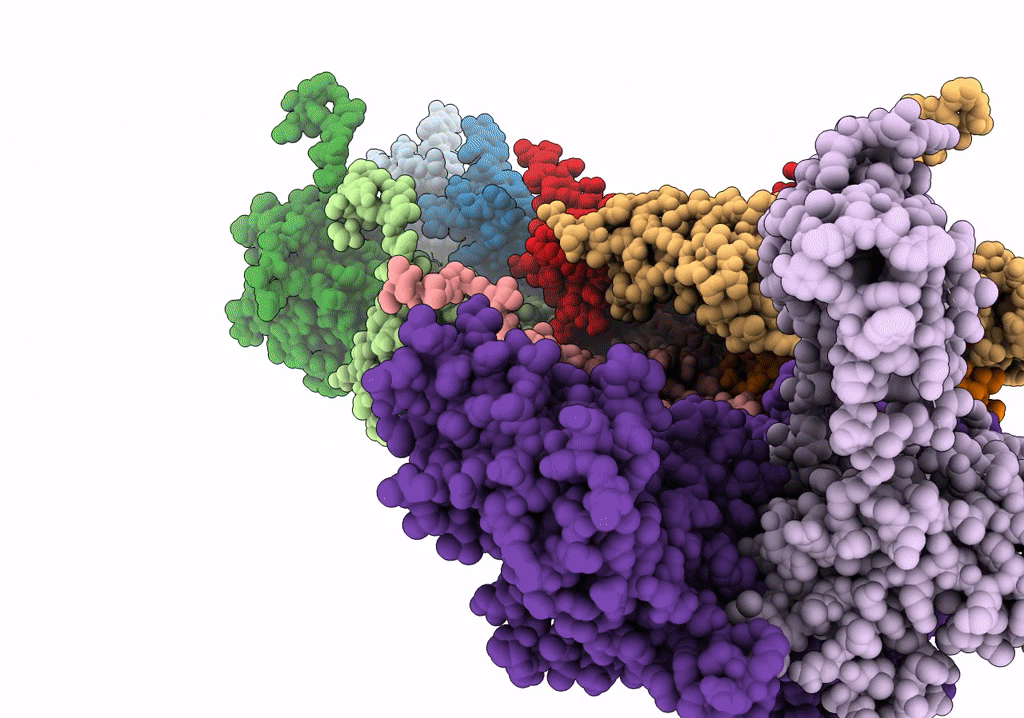
Cryo-Em Structure Of The Anti-Feeding Prophage (Afp) Baseplate In Contracted State
Organism: Serratia entomophila
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: VIRUS LIKE PARTICLE |
 |
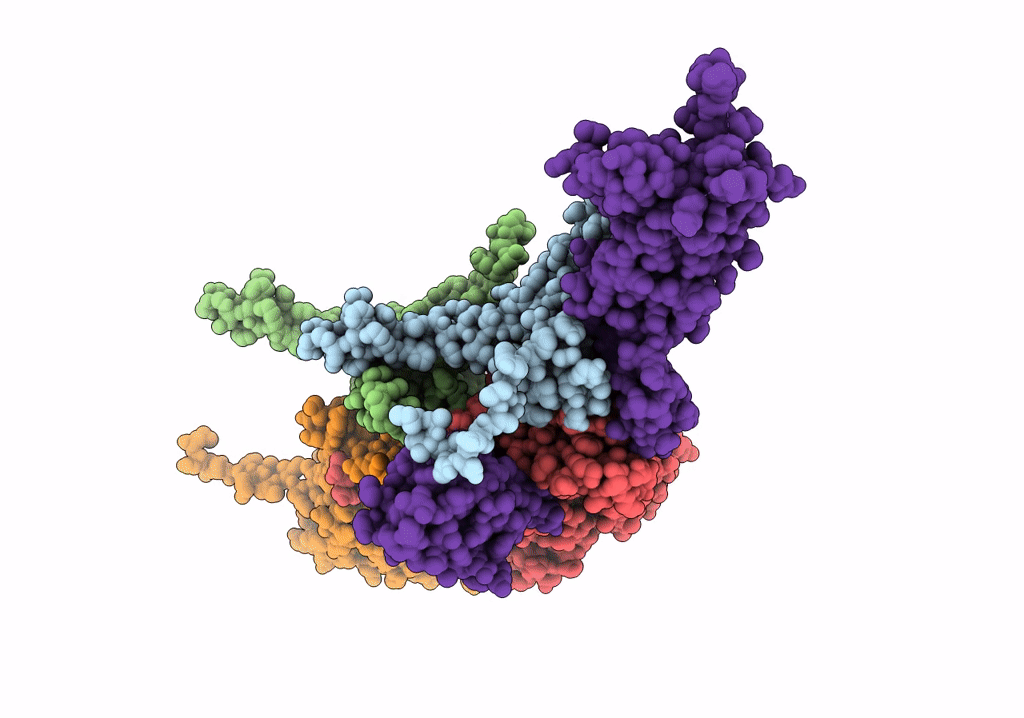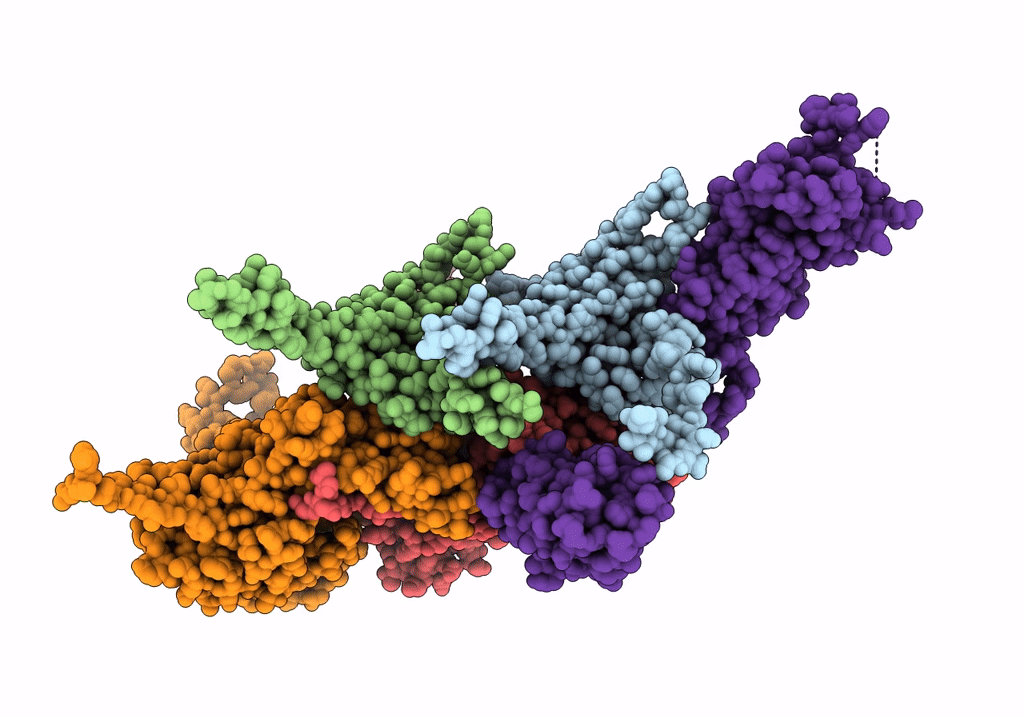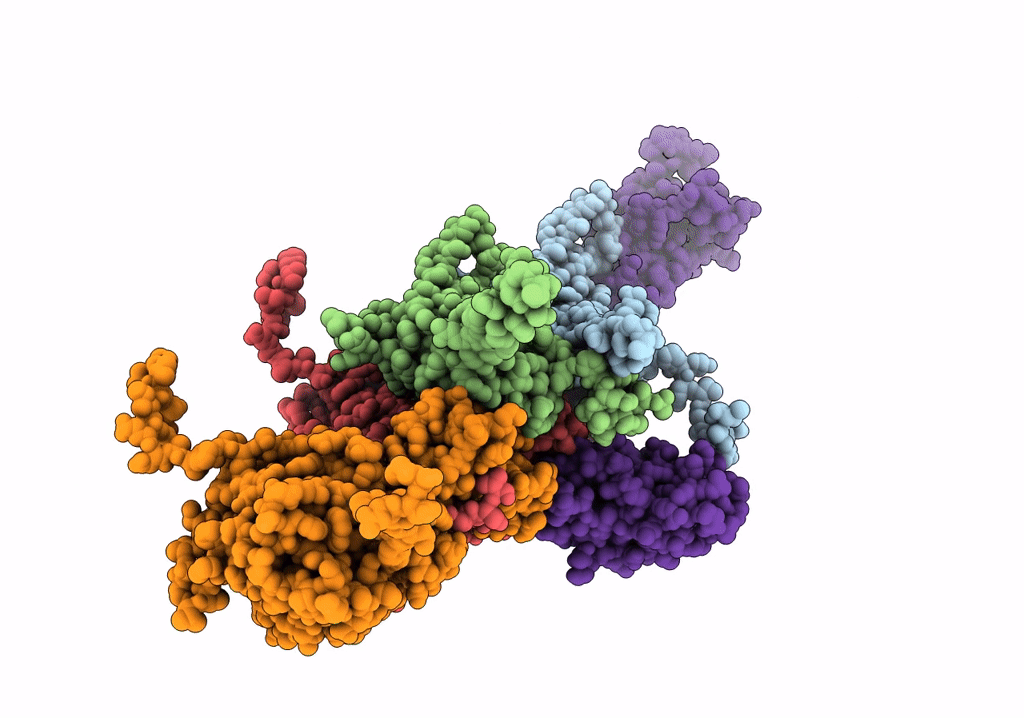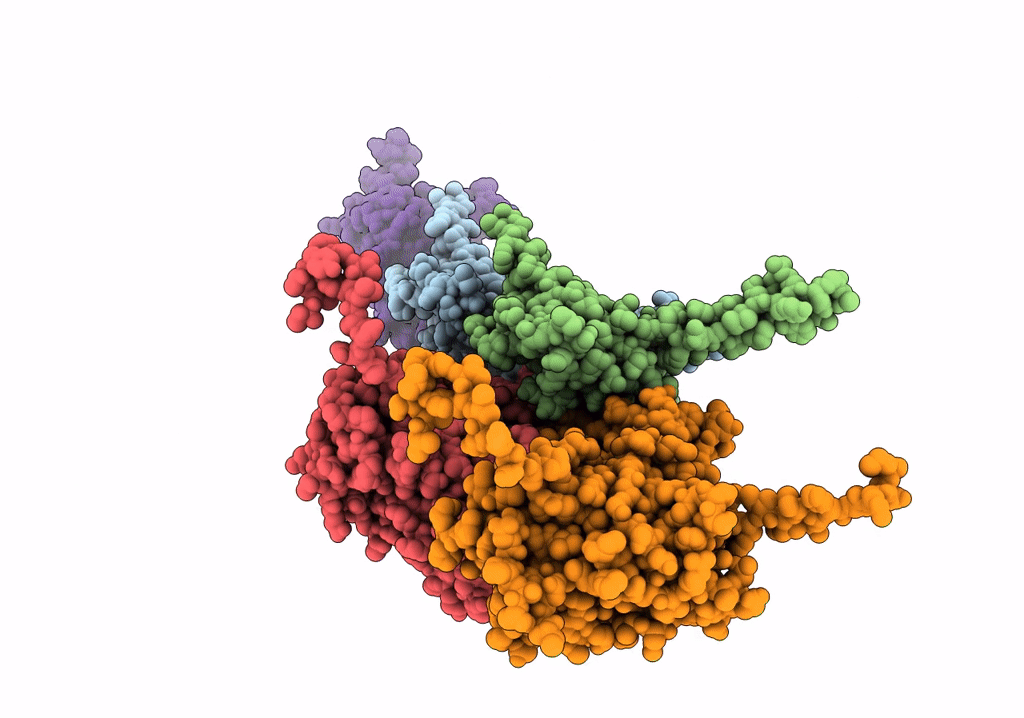
Cryo-Em Structure Of The Anti-Feeding Prophage (Afp) Baseplate, 6-Fold Symmetrised
Organism: Serratia entomophila
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The Anti-Feeding Prophage Cap (Afp Tube Terminating Cap)
Organism: Serratia entomophila
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of The Anti-Feeding Prophage (Afp) Helical Sheath In Contracted State
Organism: Serratia entomophila
Method: ELECTRON MICROSCOPY Release Date: 2019-04-17 Classification: VIRUS LIKE PARTICLE |
 |
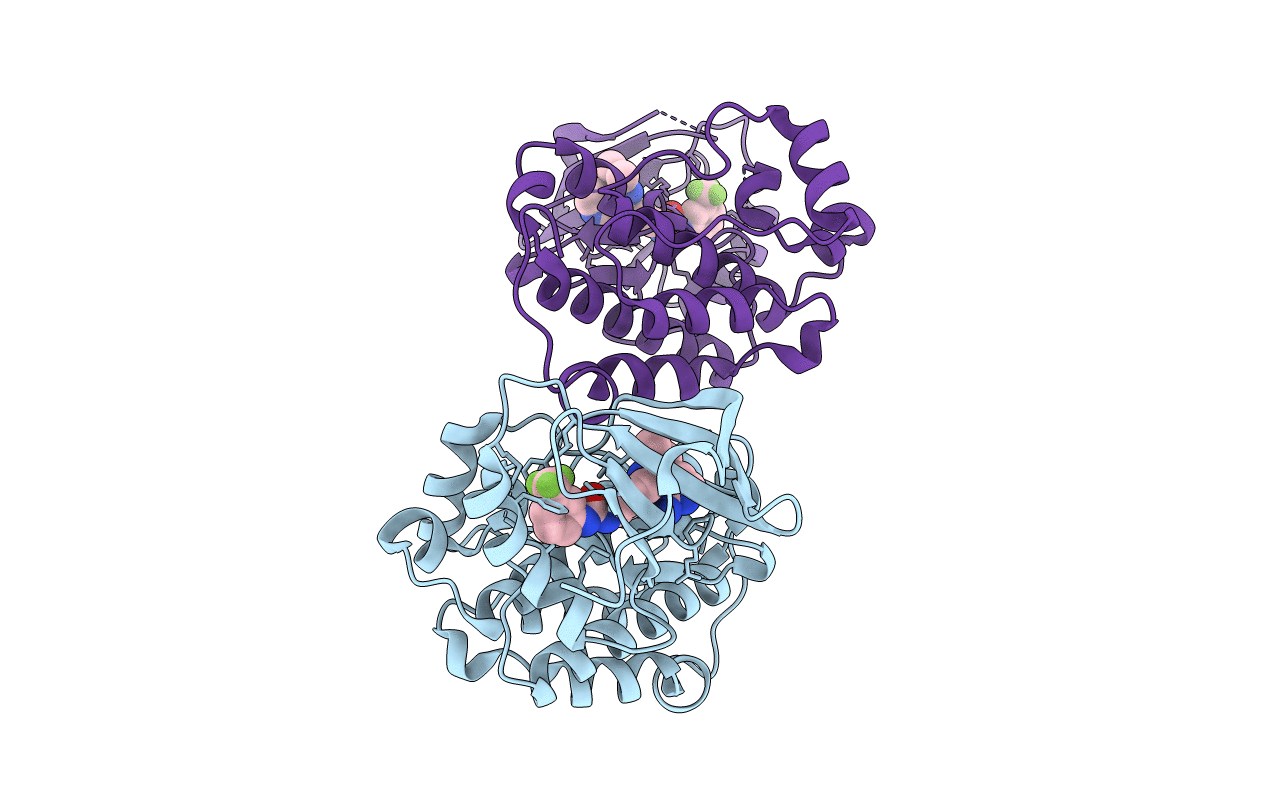
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-06-27 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2016-06-08 Classification: Transferase/Oxidoreductase Ligands: CS, NA, GOL, DTT, BME, CL |
 |
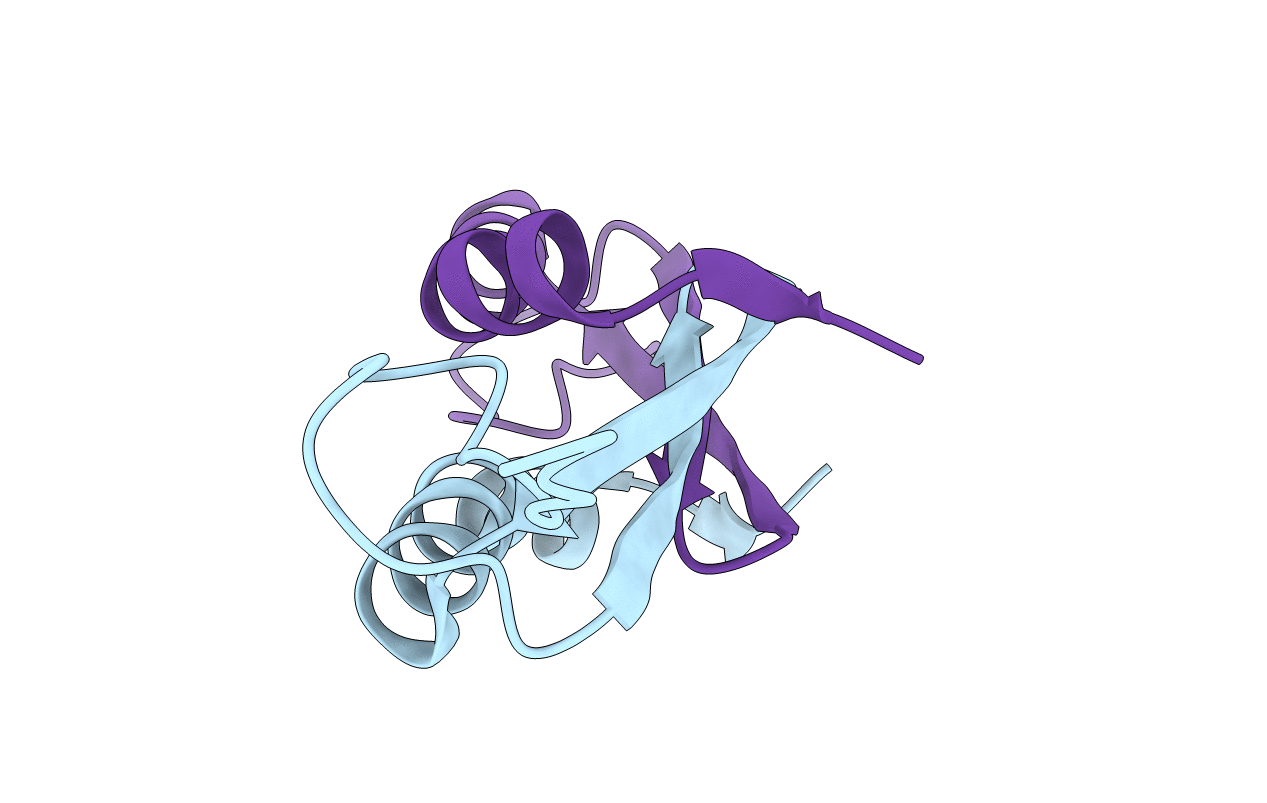
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-06-01 Classification: Transferase/Transferase Inhibitor Ligands: 6G3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-05-25 Classification: OXIDOREDUCTASE |
 |
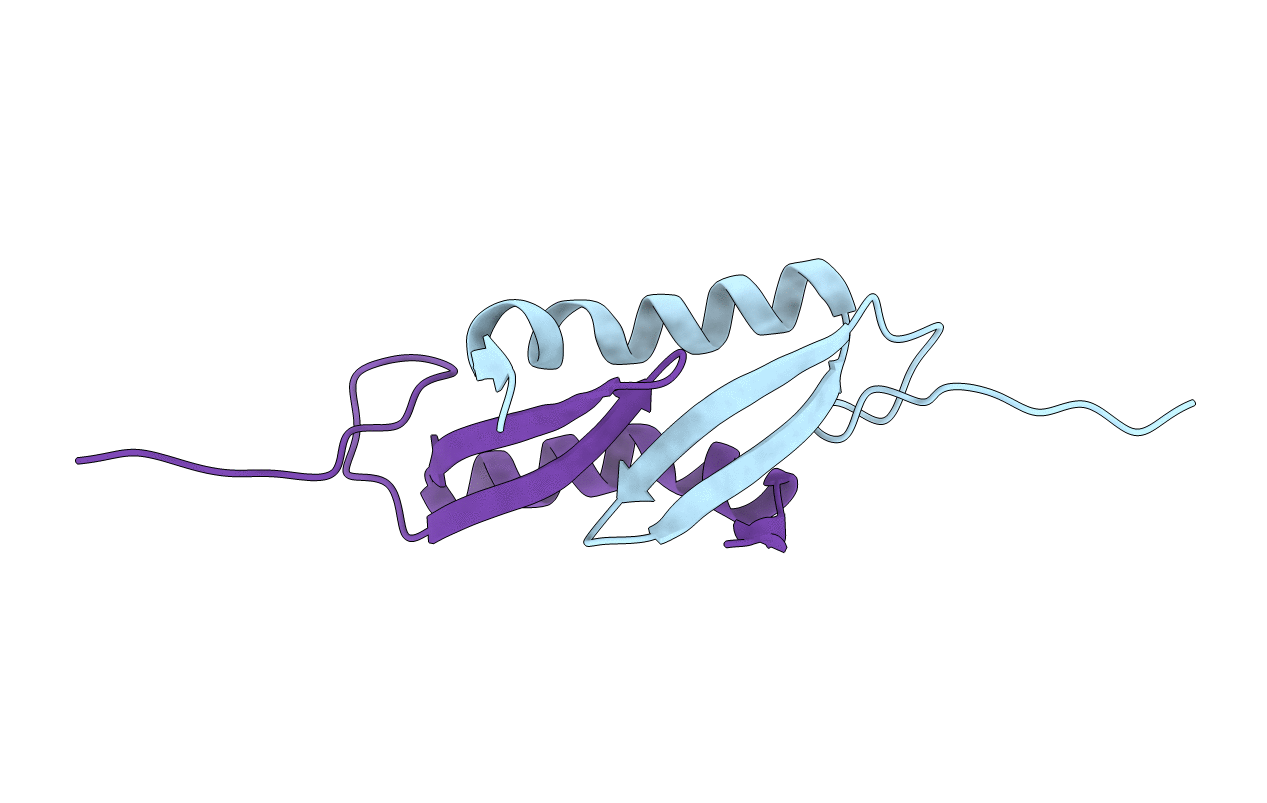
Crystal Structure Of The Dimerization Domain Of Lsr2 From Mycobacterium Tuberculosis In The P 1 21 1 Space Group
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2012-06-20 Classification: DNA BINDING PROTEIN |
 |
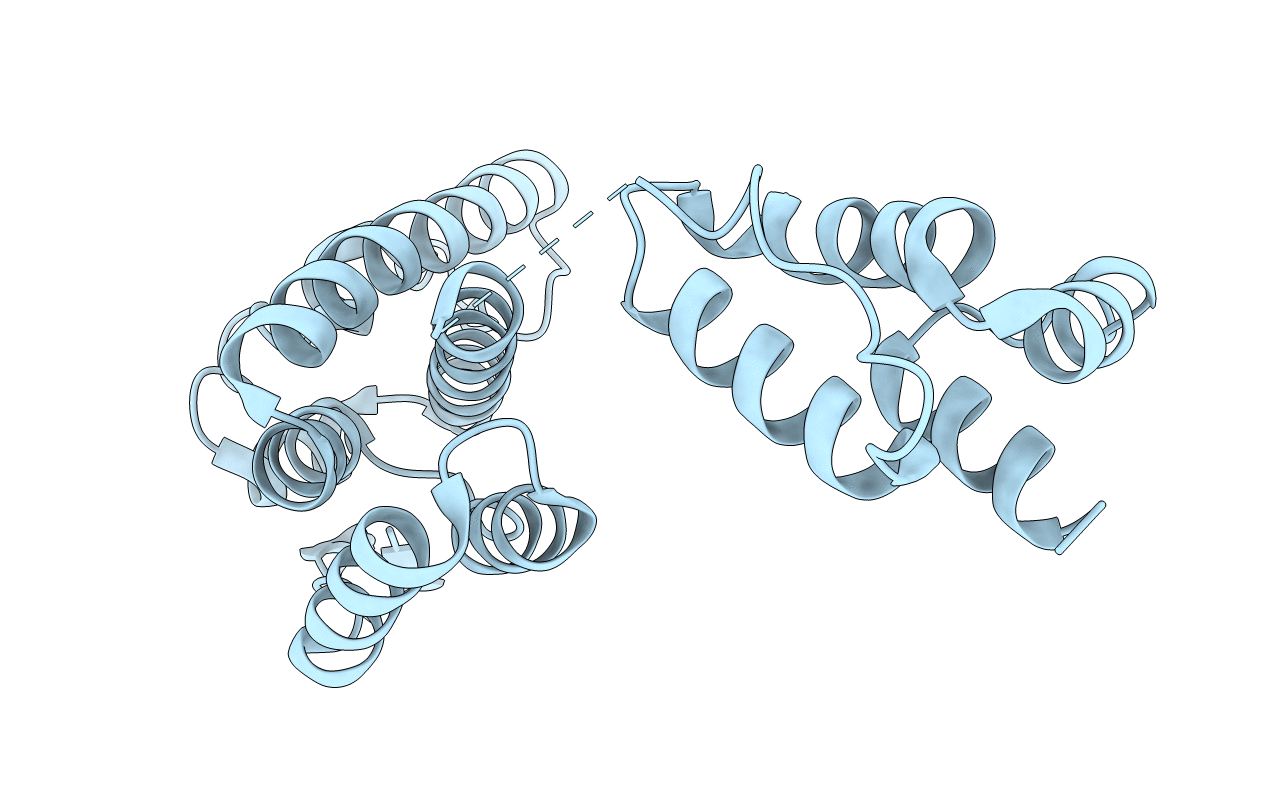
Crystal Structure Of The Dimerization Domain Of Lsr2 From Mycobacterium Tuberculosis In The P 31 2 1 Space Group
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2012-06-20 Classification: DNA BINDING PROTEIN |
 |
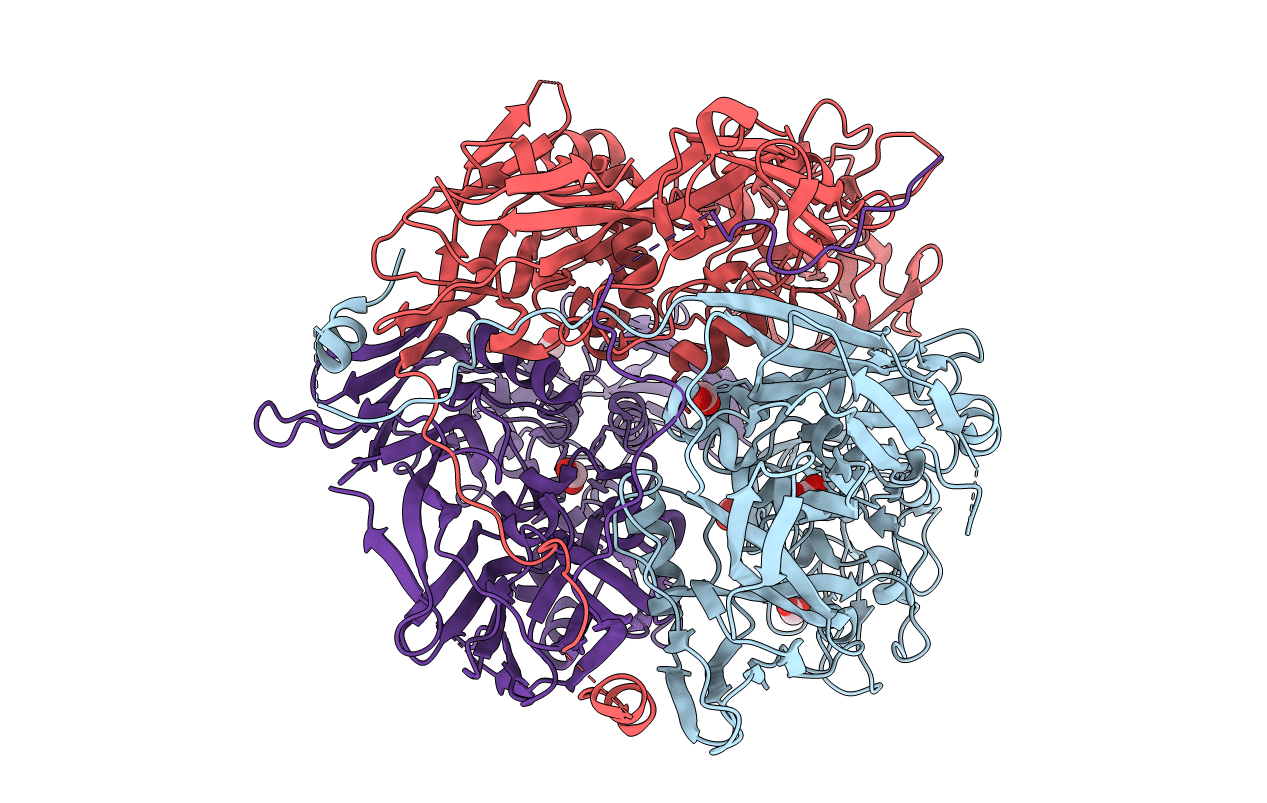
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2012-04-04 Classification: VIRAL PROTEIN |
 |
Organism: Vaccinia virus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2011-06-22 Classification: VIRAL PROTEIN Ligands: FMT |
 |
Organism: Vaccinia virus
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2011-06-22 Classification: VIRAL PROTEIN |
 |
Cryo-Em 3D Model Of The Icosahedral Particle Composed Of Rous Sarcoma Virus Capsid Protein Pentamers
Organism: Rous sarcoma virus - prague c
Method: ELECTRON MICROSCOPY Resolution:18.30 Å Release Date: 2010-05-19 Classification: VIRUS |
 |
Crystal Structure Of The C-Terminal Domain From The Rous Sarcoma Virus Capsid Protein: Mutant D179A
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-06-02 Classification: VIRAL PROTEIN Ligands: NO3 |
 |
Crystal Structure Of The C-Terminal Domain From The Rous Sarcoma Virus Capsid Protein: High Ph
Organism: Rous sarcoma virus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-06-02 Classification: VIRAL PROTEIN |

