Search Count: 16
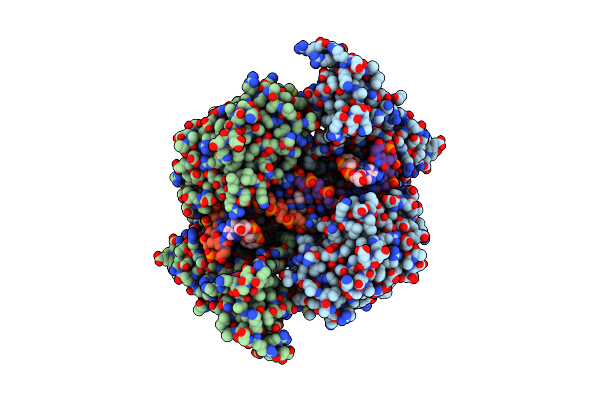 |
Ternary Complex Of A Mycobacterium Tuberculosis Dna Gyrase Core Fusion With Dna And The Inhibitor Amk32B
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-08-07 Classification: ISOMERASE Ligands: A1ID8, A1ID9 |
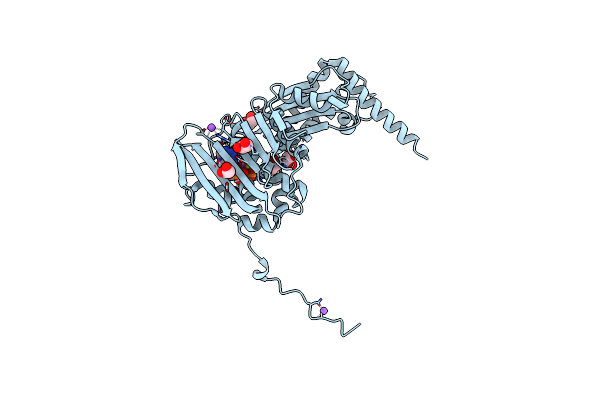 |
N-Terminal 47 Kda Fragment Of The Mycobacterium Smegmatis Dna Gyrase B Subunit Complexed With Adpnp
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2021-03-03 Classification: ISOMERASE Ligands: ANP, MG, EDO, NA, K |
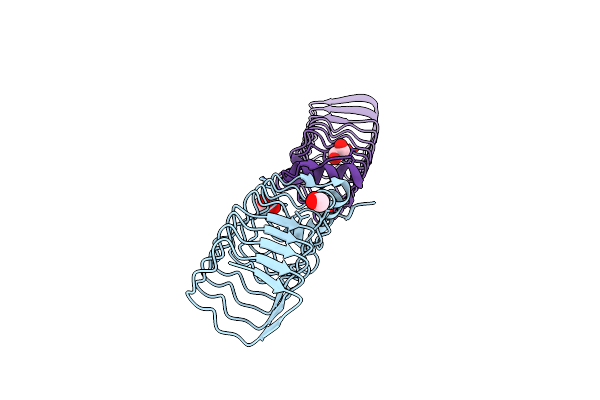 |
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-03-03 Classification: PROTEIN BINDING Ligands: EDO |
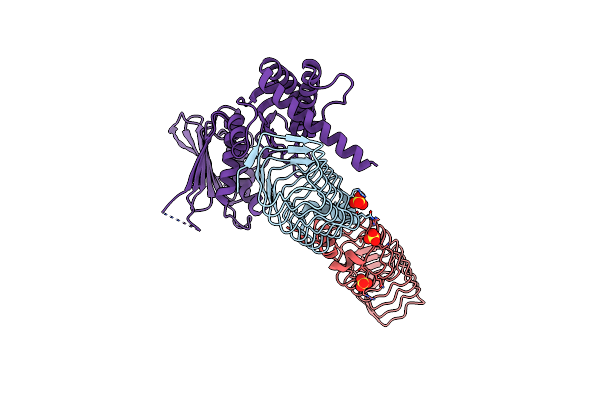 |
Complex Between A Homodimer Of Mycobacterium Smegmatis Mfpa And A Single Copy Of The N-Terminal 47 Kda Fragment Of The Mycobacterium Smegmatis Dna Gyrase B Subunit
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-03-03 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-08-12 Classification: DNA BINDING PROTEIN Ligands: NOV, ZN, EDO |
 |
Organism: Mycolicibacterium thermoresistibile
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-08-12 Classification: DNA BINDING PROTEIN Ligands: OGH, ZN, EDO |
 |
Mycobacterium Smegmatis Gyrb 22Kda Atpase Sub-Domain In Complex With Novobiocin
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-08-12 Classification: DNA BINDING PROTEIN Ligands: NOV, EDO, ACT, NA |
 |
Crystal Structure Of N-Terminal Domain Of Gyra With The Antibiotic Simocyclinone D8
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2010-12-29 Classification: ISOMERASE Ligands: MG, SM8 |
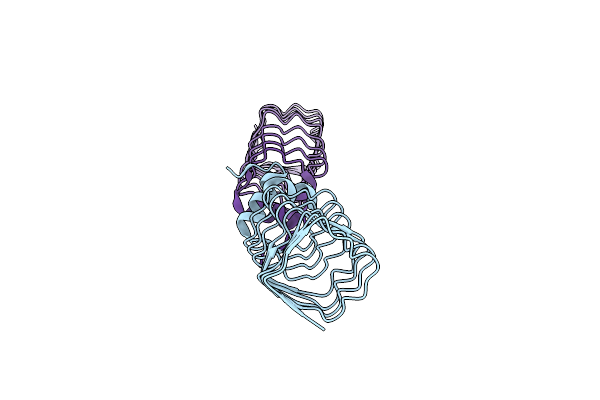 |
The Structure Of Mfpa (Rv3361C, C2 Crystal Form). The Pentapeptide Repeat Protein From Mycobacterium Tuberculosis Folds As A Right- Handed Quadrilateral Beta-Helix.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-06-07 Classification: PENTAPEPTIDE REPEAT PROTEIN |
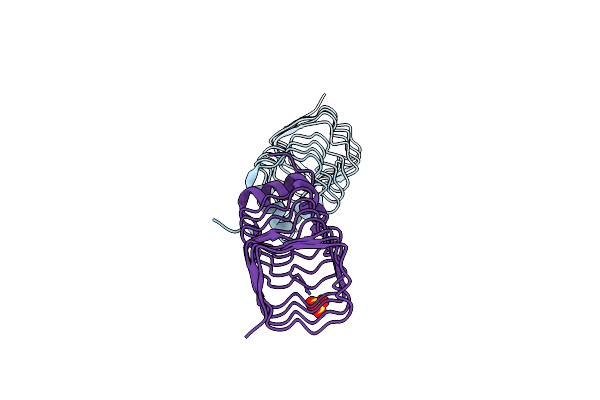 |
The Structure Of Mfpa (Rv3361C, P21 Crystal Form). The Pentapeptide Repeat Protein From Mycobacterium Tuberculosis Folds As A Right- Handed Quadrilateral Beta-Helix.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-06-07 Classification: PENTAPEPTIDE REPEAT PROTEIN Ligands: SO4 |
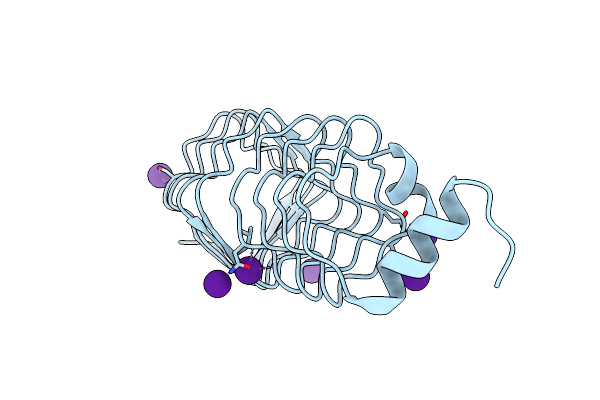 |
The Structure Of Mfpa (Rv3361C, C2221 Crystal Form). The Pentapeptide Repeat Protein From Mycobacterium Tuberculosis Folds As A Right- Handed Quadrilateral Beta-Helix.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-06-07 Classification: PENTAPEPTIDE REPEAT PROTEIN Ligands: CS |
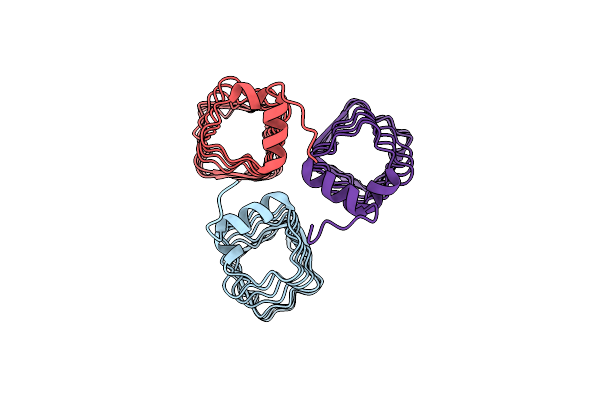 |
The Structure Of Mfpa (Rv3361C, P3221 Crystal Form). The Pentapeptide Repeat Protein From Mycobacterium Tuberculosis Folds As A Right- Handed Quadrilateral Beta-Helix.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2005-06-07 Classification: PENTAPEPTIDE REPEAT PROTEIN |
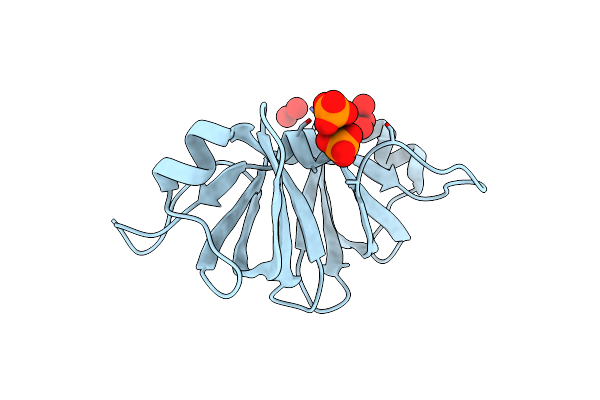 |
Two Crystal Structures Of The Cytoplasmic Molybdate-Binding Protein Modg Suggest A Novel Cooperative Binding Mechanism And Provide Insights Into Ligand-Binding Specificity. Phosphate-Grown Form With Molybdate And Phosphate Bound
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-05-11 Classification: BINDING PROTEIN Ligands: MOO, PO4 |
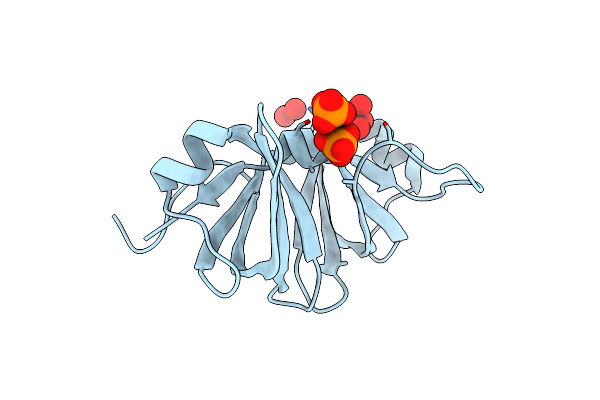 |
Two Crystal Structures Of The Cytoplasmic Molybdate-Binding Protein Modg Suggest A Novel Cooperative Binding Mechanism And Provide Insights Into Ligand-Binding Specificity. Phosphate-Grown Form With Tungstate And Phosphate Bound
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-05-11 Classification: BINDING PROTEIN Ligands: WO4, PO4 |
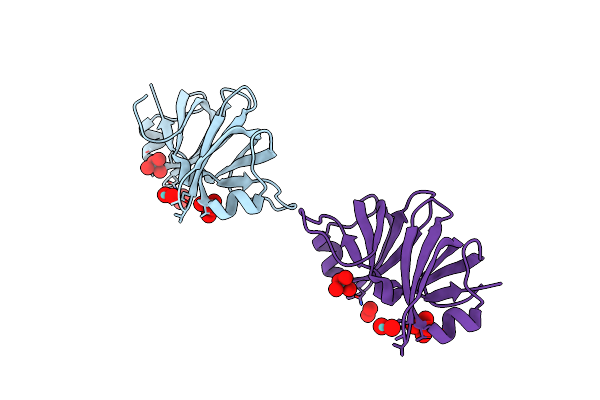 |
Two Crystal Structures Of The Cytoplasmic Molybdate-Binding Protein Modg Suggest A Novel Cooperative Binding Mechanism And Provide Insights Into Ligand-Binding Specificity. Peg-Grown Form With Molybdate Bound
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2001-05-11 Classification: BINDING PROTEIN Ligands: MOO |
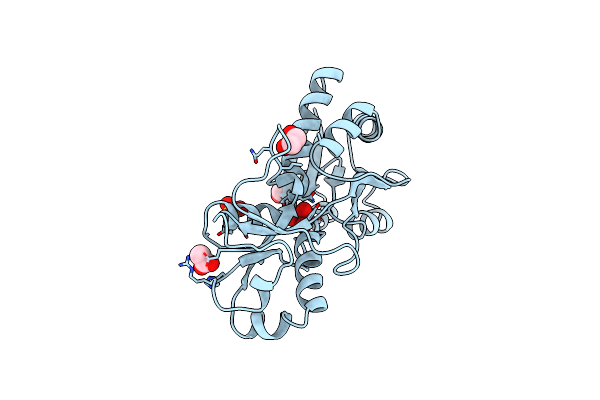 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 1998-10-14 Classification: BINDING PROTEIN Ligands: WO4, ACT, SO4, EDO |

