Search Count: 23
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YK, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YV, CL, MES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: TRANSFERASE Ligands: 5YO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: A1A2B |
 |
Pre-Clinical Characterization Of Novel Multi-Client Inhibitors Of Sec61 With Broad Anti-Tumor Activity
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: PROTEIN TRANSPORT Ligands: A1A2B |
 |
Cryoem Structure Of An Inward-Facing Melbst At A Na(+)-Bound And Sugar Low-Affinity Conformation
Organism: Salmonella enterica subsp. enterica serovar typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Human Mitochondrial Dna Polymerase Gamma Ternary Complex With Gt Basepair In Replication Conformer
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Human Mitochondrial Dna Polymerase Gamma Ternary Complex With Gt Basepair In Intermediate Conformer
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSFERASE/DNA Ligands: DCP, CA |
 |
Human Mitochondrial Dna Polymerase Gamma Ternary Complex With Gt Basepair In Editing Conformer (Composite)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSFERASE/DNA Ligands: CA, DCP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-07-27 Classification: TRANSFERASE Ligands: 5YS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2016-08-24 Classification: SIGNALING PROTEIN Ligands: PGE, P33, PEG, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2016-08-24 Classification: LIPID BINDING PROTEIN Ligands: 6RQ, PEG, P33 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2016-08-24 Classification: LIPID BINDING PROTEIN Ligands: PEG, CHD |
 |
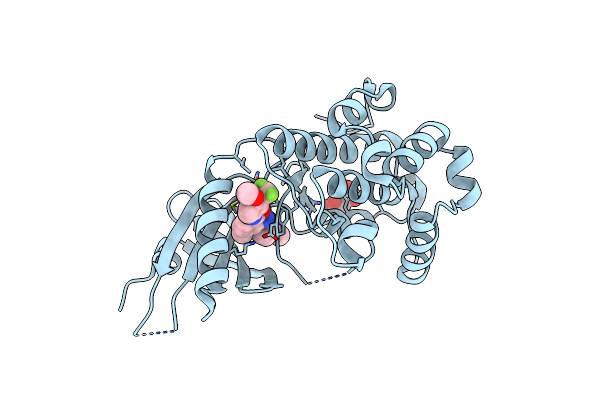
Co-Crystal Structure Of Perk Bound To N-{5-[(6,7-Dimethoxyquinolin-4-Yl)Oxy]Pyridin-2-Yl}-1-Methyl-3-Oxo-2-Phenyl-5-(Pyridin-4-Yl)-2,3-Dihydro-1H-Pyrazole-4-Carboxamide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-02-04 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3Z2, SO4 |
 |
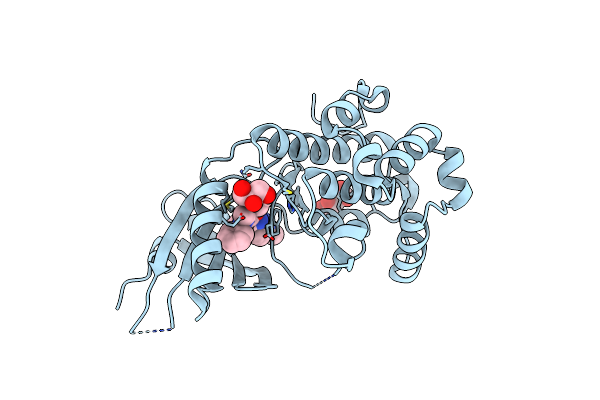
Co-Crystal Structure Of Perk With 2-Amino-N-[4-Methoxy-3-(Trifluoromethyl)Phenyl]-4-Methyl-3-[2-(Methylamino)Quinazolin-6-Yl]Benzamide Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3Z1, TLA |
 |
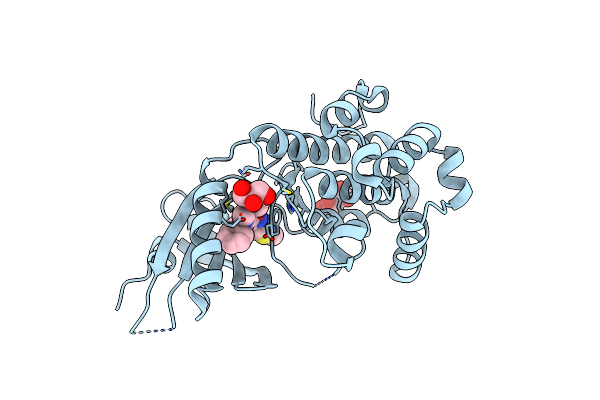
Co-Crystal Structure Of Perk Bound To 4-{2-Amino-3-[5-Fluoro-2-(Methylamino)Quinazolin-6-Yl]-4-Methylbenzoyl}-1-Methyl-2,5-Diphenyl-1,2-Dihydro-3H-Pyrazol-3-One Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3Z3, TLA, GOL |
 |
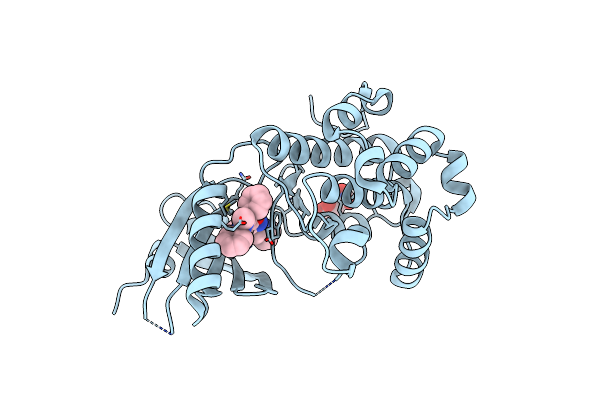
Co-Crystal Structure Of Perk Bound To 4-{2-Amino-4-Methyl-3-[2-(Methylamino)-1,3-Benzothiazol-6-Yl]Benzoyl}-1-Methyl-2,5-Diphenyl-1,2-Dihydro-3H-Pyrazol-3-One Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3Z4, TLA, GOL |
 |
Co-Crystal Structure Of Perk Bound To 4-[2-Amino-4-Methyl-3-(2-Methylquinolin-6-Yl)Benzoyl]-1-Methyl-2,5-Diphenyl-1,2-Dihydro-3H-Pyrazol-3-One Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2015-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3Z5, TLA |
 |
Co-Crystal Structure Of Perk Bound To 1-[5-(4-Amino-2,7-Dimethyl-7H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)-2,3-Dihydro-1H-Indol-1-Yl]-2-[3-Fluoro-5-(Trifluoromethyl)Phenyl]Ethanone Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 3Z6 |

