Search Count: 39
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: CA, EDO |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: T83, CA, EDO, PG4, ACT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: 07L, CA, ACT |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-01-11 Classification: PLANT PROTEIN Ligands: COA, GOL, CA |
 |
Dr3 In Complex With Aspergillus Nidulans Nad-Dependent Histone Deacetylase Hst4 Peptide
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2021-08-18 Classification: IMMUNE SYSTEM Ligands: EDO, NAG |
 |
Crystal Structure Of A Non-Hemolytic Pneumolysin From Streptococcus Pneumoniae Strain St306
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-09-09 Classification: TOXIN Ligands: GOL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-09-18 Classification: OXIDOREDUCTASE Ligands: DAO, FE, GOL |
 |
Crystal Structure Of Coleus Blumei Rosmarinic Acid Synthase (Ras) In Complex With 4-Coumaroyl-(R)-3-(4-Hydroxyphenyl)Lactate
Organism: Plectranthus scutellarioides
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2019-09-11 Classification: TRANSFERASE Ligands: JUV |
 |
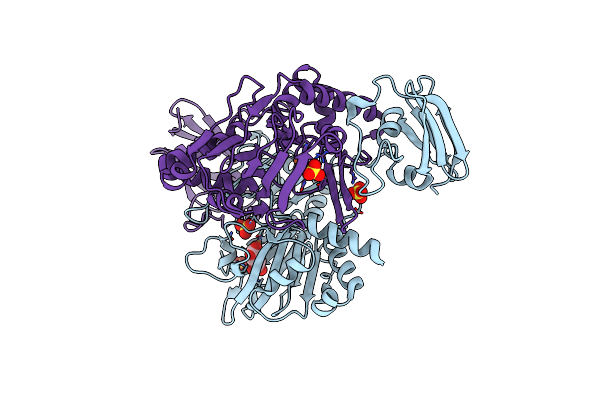
X-Ray Crystal Structure Of Tmpb, (R)-1-Hydroxy-2-Trimethylaminoethylphosphonate Oxygenase, With (R)-1-Hydroxy-2-Trimethylaminoethylphosphonate
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE, FE2, KVP |
 |
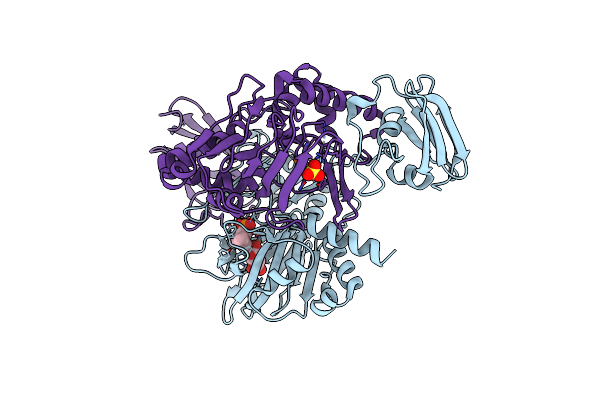
X-Ray Crystal Structure Of Tmpa, 2-Trimethylaminoethylphosphonate Hydroxylase, With Fe And 2Og
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE2, AKG, SO4 |
 |
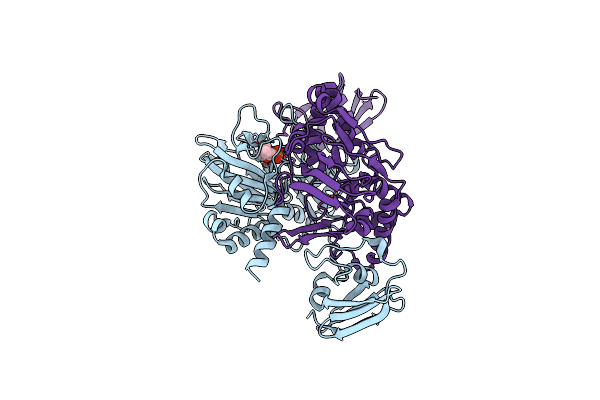
X-Ray Crystal Structure Of Tmpa, 2-Trimethylaminoethylphosphonate Hydroxylase, With Fe, 2Og, And 2-Trimethylaminoethylphosphonate
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE2, AKG, KVV, SO4 |
 |
X-Ray Crystal Structure Of Tmpa, 2-Trimethylaminoethylphosphonate Hydroxylase, With Fe, 2Og, And (R)-1-Hydroxy-2-Trimethylaminoethylphosphonate
Organism: Leisingera caerulea
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE Ligands: FE2, KVP |
 |
Crystal Structure Of Sznf From Streptomyces Achromogenes Var. Streptozoticus Nrrl 2697 With A Bound N(Delta)-Hydroxy-N(Omega)-Methyl-L-Arginine Intermediate
Organism: Streptomyces achromogenes subsp. streptozoticus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2019-02-06 Classification: OXIDOREDUCTASE Ligands: J9Y, FE |
 |
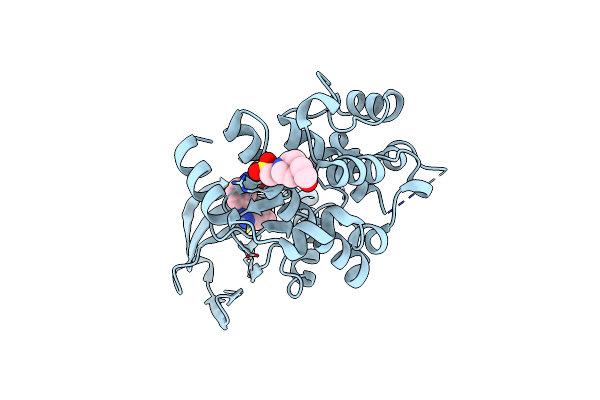
Crystal Structure Of Semet Sznf From Streptomyces Achromogenes Var. Streptozoticus Nrrl 2697
Organism: Streptomyces achromogenes subsp. streptozoticus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-02-06 Classification: OXIDOREDUCTASE Ligands: FE, GOL, SO4, IPA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2019-01-02 Classification: TRANSFERASE Ligands: 95U, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-01-02 Classification: TRANSFERASE Ligands: 95U |
 |
X-Ray Crystal Structure Of Vioc Bound To Fe(Ii), D-Arginine, And 2-Oxoglutarate
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-11-21 Classification: OXIDOREDUCTASE Ligands: AKG, FE2, DAR |
 |
X-Ray Crystal Structure Of Vioc Bound To Fe(Ii), 2-Oxo-5-Guanidinopentanoic Acid, And Succinate
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-11-21 Classification: OXIDOREDUCTASE Ligands: JX7, SIN, FE2 |
 |
X-Ray Crystal Structure Of Napi L-Arginine Desaturase Bound To Fe(Ii), L-Arginine, And Acetate
Organism: Streptomyces lusitanus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: FE2, ARG, ACT |
 |
X-Ray Crystal Structure Of Vioc Bound To Fe(Ii), L-Homoarginine, And 2-Oxoglutarate
Organism: Streptomyces vinaceus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-16 Classification: OXIDOREDUCTASE Ligands: AKG, HRG, FE2 |

