Search Count: 43
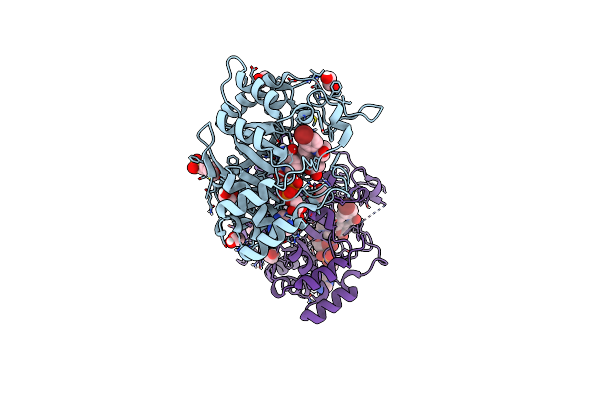 |
Structure Of Aldo-Keto Reductase 1C3 (Akr1C3) In Complex With An Inhibitor Meds765
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-08-21 Classification: OXIDOREDUCTASE Ligands: NAP, A1ICG, EDO, MES |
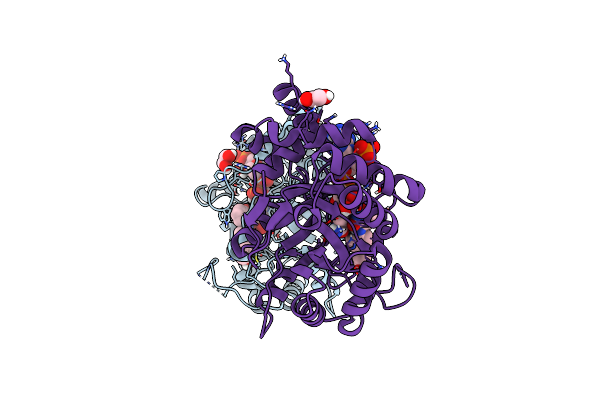 |
Structure Of Aldo-Keto Reductase 1C3 (Akr1C3) In Complex With An Inhibitor M689, With The 3-Hydroxy-Benzoisoxazole Moiety. Resolution 2.0A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-06 Classification: OXIDOREDUCTASE Ligands: NAP, YMC, EDO |
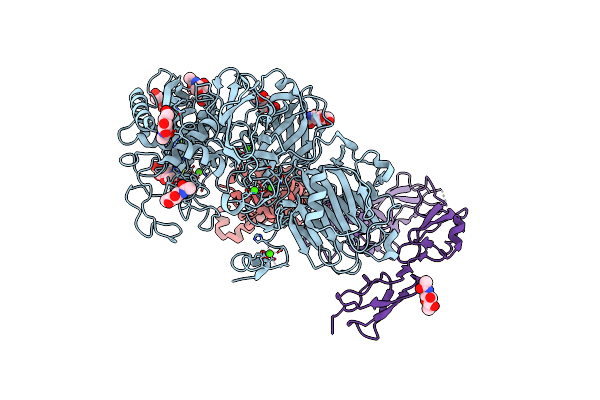 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: HYDROLASE Ligands: NAG, ZN, CA |
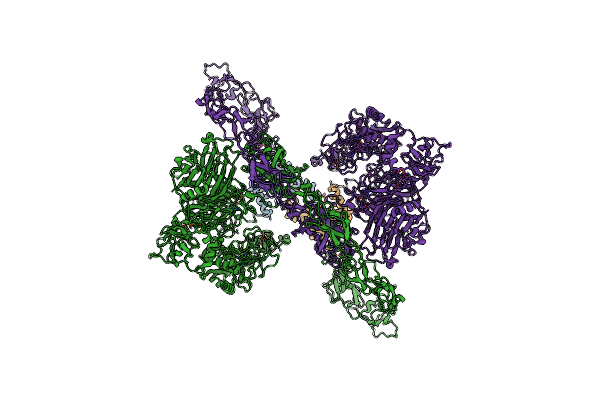 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, CA |
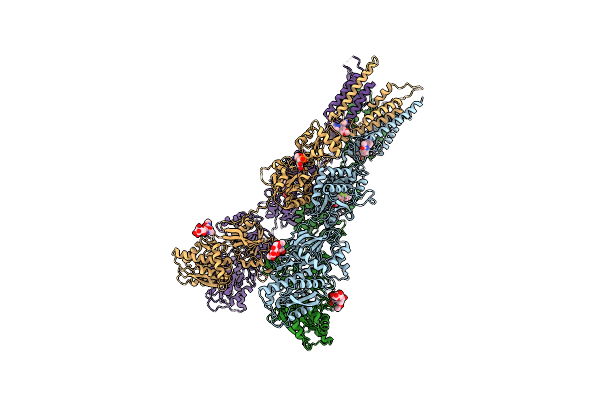 |
Structure Of Glua2Cryst In Complex The Antagonist Zk200775 And The Negative Allosteric Modulator Gyki53655 At 4.65 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:4.65 Å Release Date: 2020-06-24 Classification: MEMBRANE PROTEIN Ligands: GYK, ZK1 |
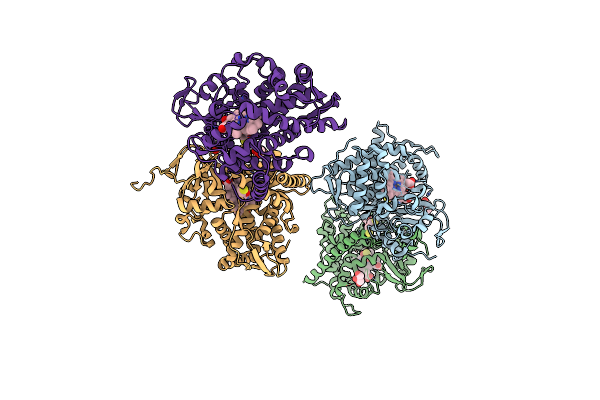 |
Structure Of Cytochrome P450 Bm3 M11 Mutant In Complex With Dtt At Resolution 2.16A
Organism: Bacillus megaterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2019-06-05 Classification: OXIDOREDUCTASE Ligands: HEM, DTT, CL, GOL, PEG |
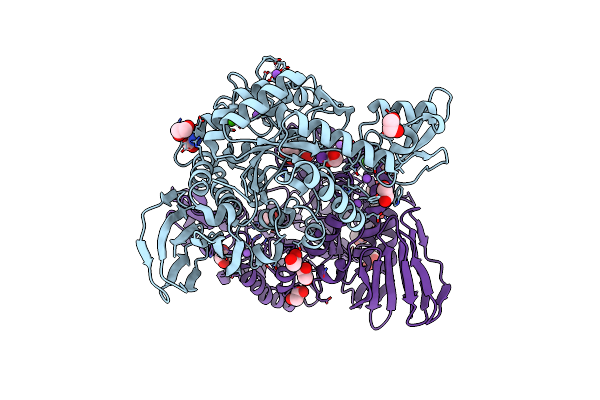 |
Functional Characterization And Crystal Structure Of Thermostable Amylase From Thermotoga Petrophila, Reveals High Thermostability And An Archaic Form Of Dimerization
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-07-19 Classification: HYDROLASE |
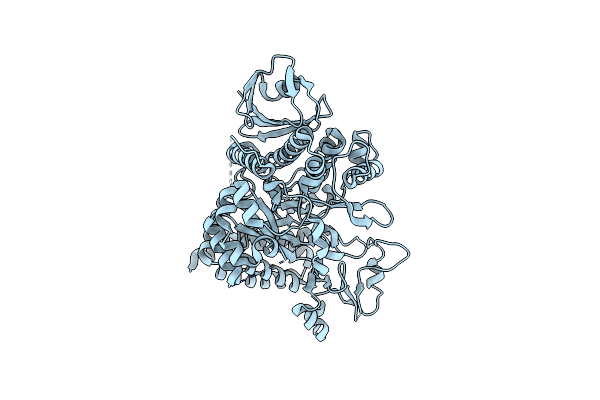 |
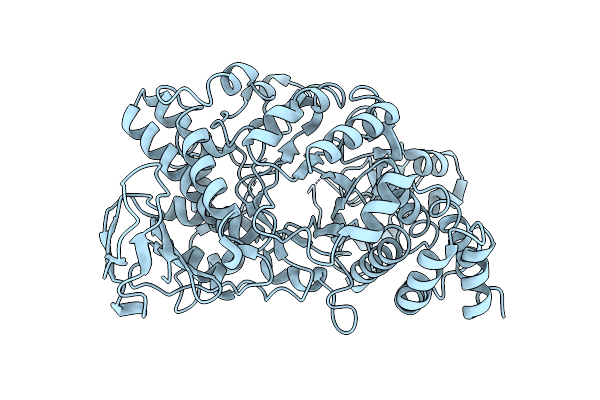
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2013-09-11 Classification: TRANSFERASE |
 |
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-11-24 Classification: HYDROLASE |
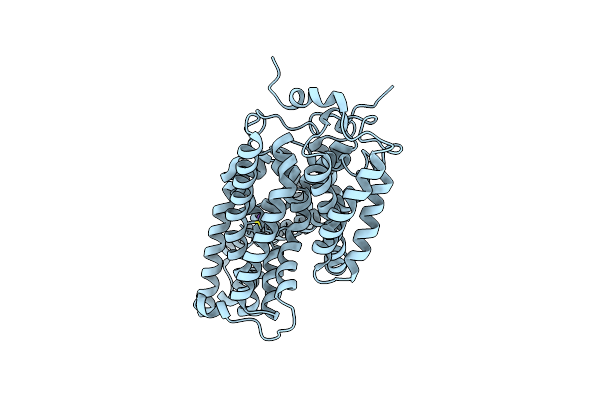 |
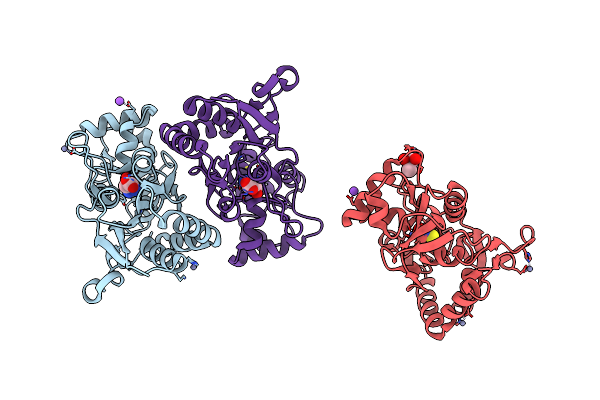
Organism: Microbacterium liquefaciens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2008-10-28 Classification: MEMBRANE PROTEIN |
 |
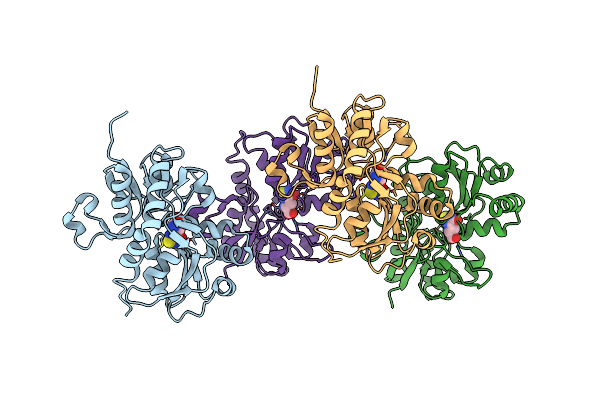
Structure Of The Ligand-Binding Core Of Glur2 In Complex With The Agonist (S)-Tdpa At 2.25 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2008-10-28 Classification: MEMBRANE PROTEIN Ligands: S2P, ZN, NA, CL, CAC |
 |
Structure Of The Ligand-Binding Core Of Glur2 In Complex With The Agonist (R)-Tdpa At 1.95 A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-10-14 Classification: MEMBRANE PROTEIN Ligands: R2P |
 |
Structure Of A Seleno-Methionyl Derivative Of Wild Type Arabinofuranosidase From Thermobacillus Xylanilyticus
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-07-01 Classification: HYDROLASE Ligands: PO4 |
 |
Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide
Organism: Thermobacillus xylanilyticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-07-01 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of Phosphate Preplasmic Binding Protein Psts From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN,IMMUNE SYSTEM Ligands: PO4 |
 |
Crystal Structure Of Y.Pestis Oligo Peptide Binding Protein Oppa With Tri-Lysine Ligand
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-30 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2007-09-11 Classification: TRANSPORT PROTEIN |
 |
E232Q Mutant Of Sucrose Phosphorylase From Bifidobacterium Adolescentis In Complex With Sucrose
Organism: Bifidobacterium adolescentis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-09-26 Classification: TRANSFERASE |
 |
Sucrose Phosphorylase From Bifidobacterium Adolescentis Reacted With Sucrose
Organism: Bifidobacterium adolescentis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-26 Classification: TRANSFERASE Ligands: BGC |
 |
The Structure Of A Mixed Glur2 Ligand-Binding Core Dimer In Complex With (S)-Glutamate And The Antagonist (S)-Ns1209
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2006-06-06 Classification: ION CHANNEL Ligands: SO4, M1L, GLU |

