Search Count: 7
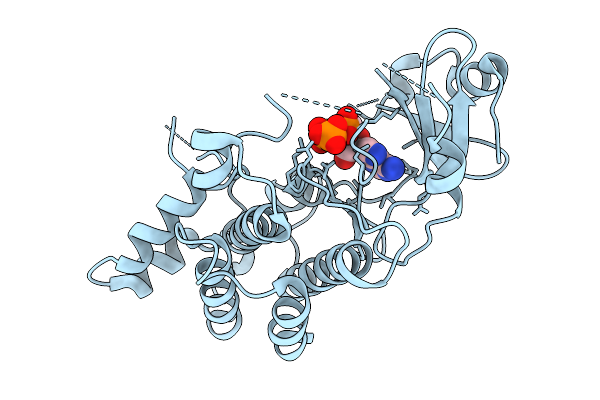 |
Structural Basis Of Signal Activation And Transduction By Chitin Elicitor Receptor Kinase 1 In Oryza Sativa
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: ANP |
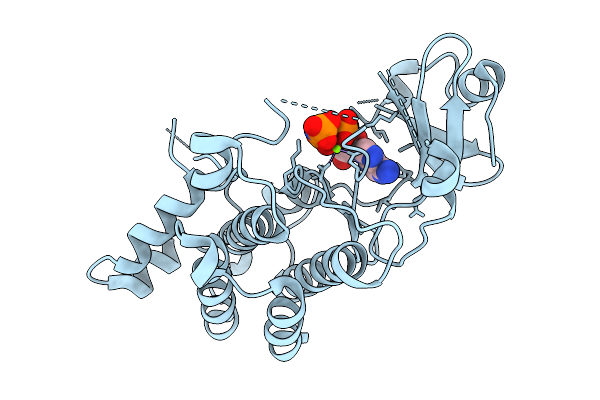 |
Structural Basis Of Signal Activation And Transduction By Chitin Elicitor Receptor Kinase 1 In Oryza Sativa
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: MG, ANP |
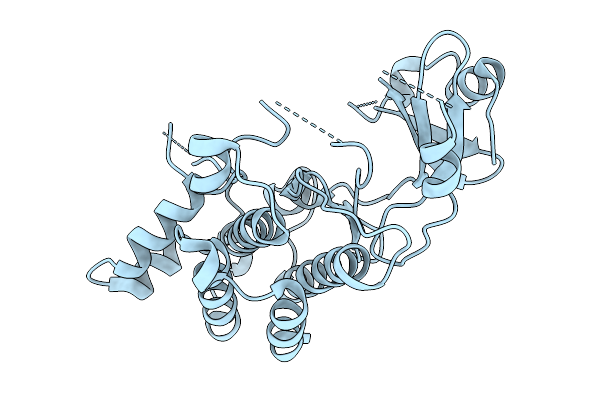 |
Structural Basis Of Signal Activation And Transduction By Chitin Elicitor Receptor Kinase 1 In Oryza Sativa
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-28 Classification: TRANSFERASE |
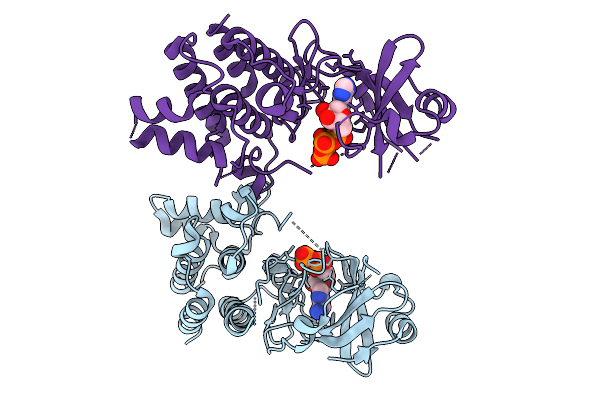 |
Structural Basis Of Signal Activation And Transduction By Chitin Elicitor Receptor Kinase 1 In Oryza Sativa
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: ATP, MG |
 |
Structural Basis Of Signal Activation And Transduction By Chitin Elicitor Receptor Kinase 1 In Oryza Sativa
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: ADP, MG |
 |
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-11-02 Classification: TRANSFERASE |
 |
Organism: Betacoronavirus england 1
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2015-08-19 Classification: HYDROLASE Ligands: ZN |

