Search Count: 8
 |
Structure Of Human Serum Albumin In Complex With Aristolochic Acid Ii At 1.9 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: LIPID BINDING PROTEIN Ligands: MYR, EDO, GOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: LIPID BINDING PROTEIN Ligands: MYR, EDO |
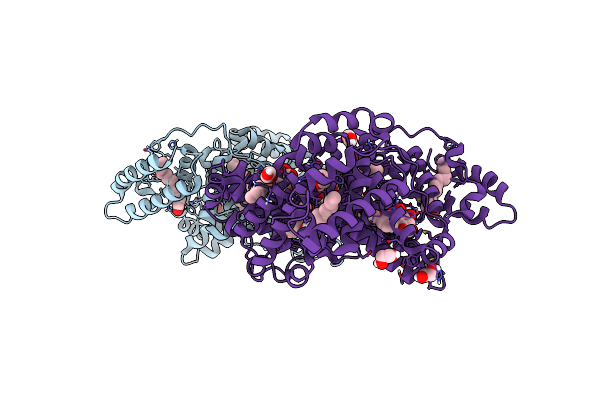 |
Structure Of Human Serum Albumin In Complex With Aristolochic Acid At 1.9 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: MYR, EDO, GOQ |
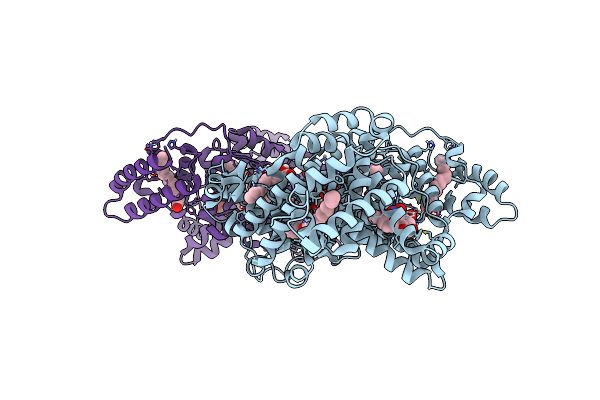 |
Structure Of Human Serum Albumin In Complex With Aristolochic Acid I At 1.9 A Resolution - Optimized
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: MYR, EDO, GOQ |
 |
Crystal Structure Of A Circular Permutation And Computationally Designed Pro-Enzyme Of Carboxypeptidase G2
Organism: Pseudomonas sp. (strain rs-16), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2021-04-14 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
 |
 |
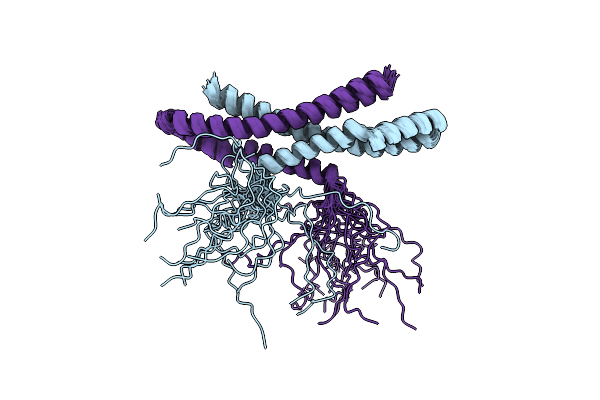
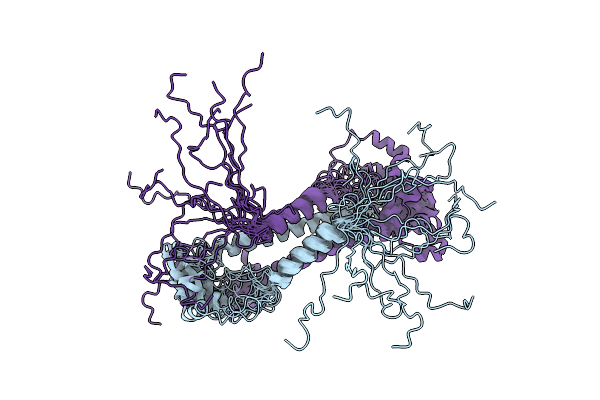
Arginine 116 Is Essential For Nucleic Acid Recognition By The Fingers Domain Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-01-27 Classification: TRANSFERASE |

