Search Count: 24
 |
Crystal Structure Of L-Ribulose 3-Epimerase From Arthrobacter Globiformis M30
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: MG, PEG |
 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-12 Classification: ISOMERASE Ligands: PSJ, FUD, MG, NA, PEG |
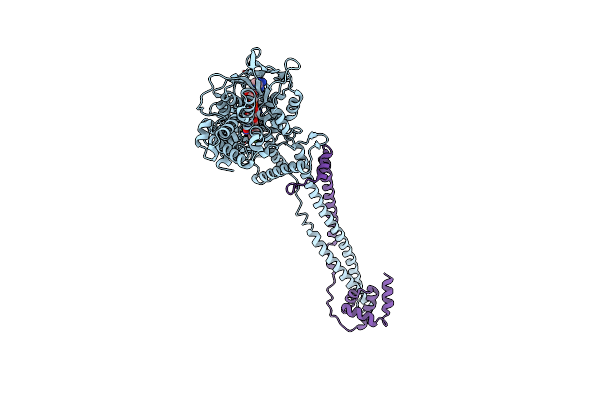 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: A1IG2 |
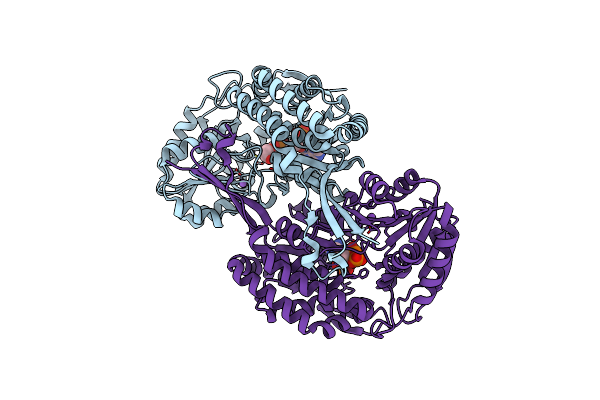 |
Crystal Structure Of Thermostable Acetaldehyde Dehydrogenase From Hyperthermophilic Archaeon Sulfolobus Tokodaii
Organism: Sulfurisphaera tokodaii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: NA7, NA, ATR |
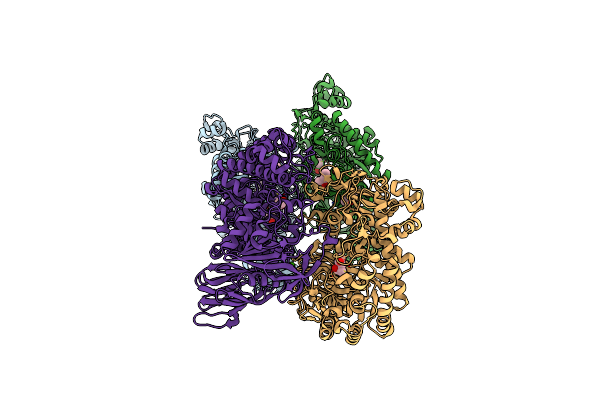 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-25 Classification: HYDROLASE Ligands: CA, MPD |
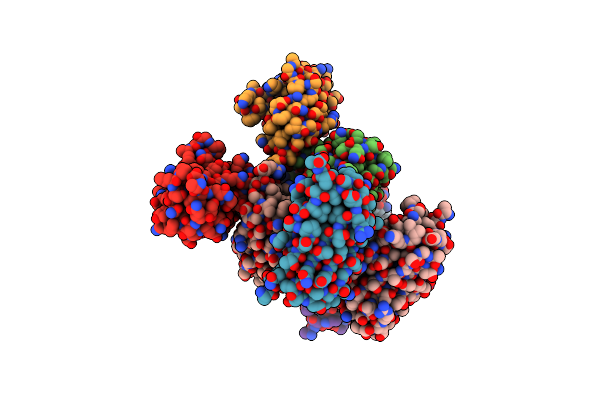 |
Structure Of Monomeric Mutant Of Rei Immunoglobulin Light Chain Variable Domain Crystallized At Ph 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2017-08-02 Classification: IMMUNE SYSTEM |
 |
Structure Of Monomeric Mutant Of Rei Immunoglobulin Light Chain Variable Domain Crystallized At Ph 8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-08-02 Classification: IMMUNE SYSTEM |
 |
Organism: Talaromyces cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-05-31 Classification: HYDROLASE Ligands: ACT, CA, ZN, PEG |
 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: PO4 |
 |
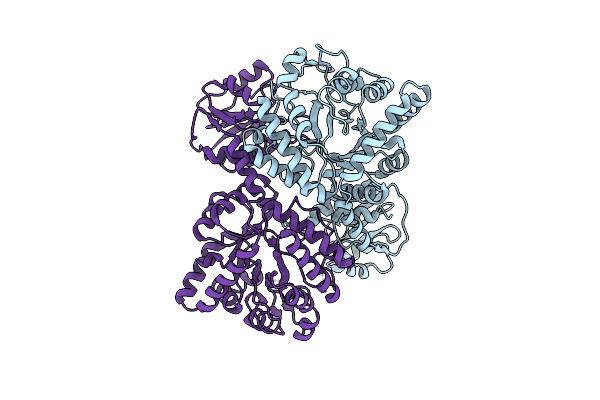
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: GCS, PEG, PPI, B3P |
 |
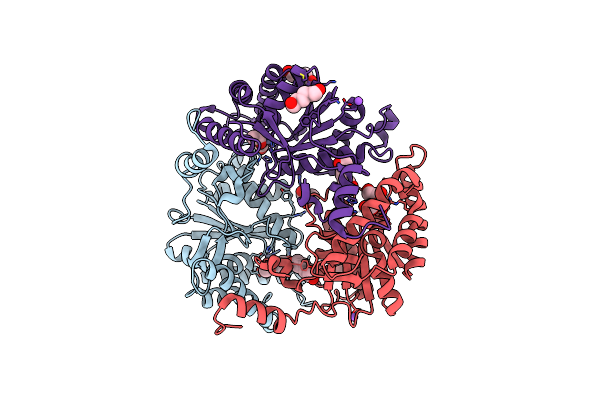
Crystal Structure Of The Beta-N-Acetylglucosaminidase From Thermotoga Maritima
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2014-12-24 Classification: HYDROLASE |
 |
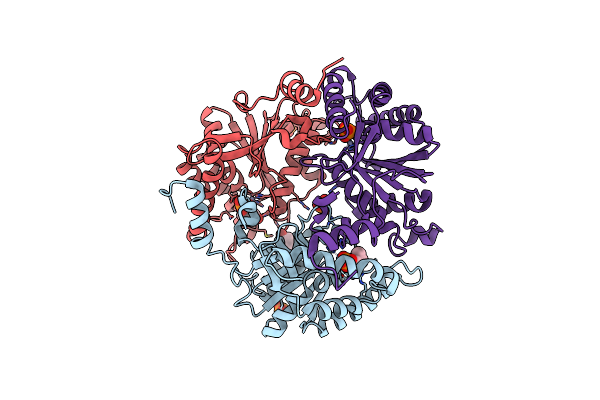
Crystal Structure Of Diacetylchitobiose Deacetylase From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-05-07 Classification: HYDROLASE Ligands: ZN, GOL, ACY, NA, HEZ |
 |
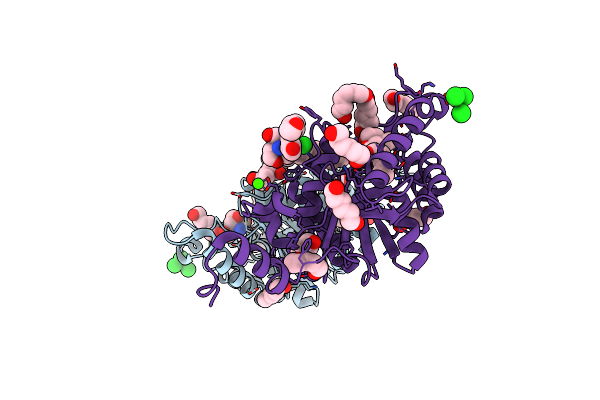
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-07 Classification: HYDROLASE Ligands: ZN, PO4, GOL |
 |
N,N'-Diacetylchitobiose Deacetylase (Se-Derivative) From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2014-05-07 Classification: HYDROLASE Ligands: CD, CA, CL, TAM, HEZ, HE2 |
 |
Organism: Pyrococcus furiosus
Method: SOLUTION NMR Release Date: 2014-04-23 Classification: HYDROLASE |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2012-06-27 Classification: TRANSFERASE Ligands: ZN, GOL |
 |
Crystal Structure Of Catalytic Domain Of Hyperthermophilic Chitinase From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-02-06 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Solution Structure Of The Chitin-Binding Domain Of Hyperthermophilic Chitinase From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: SOLUTION NMR Release Date: 2006-07-18 Classification: HYDROLASE |
 |
Crystal Structure Of Chitin Biding Domain Of Chitinase From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-07-11 Classification: HYDROLASE |
 |
The Solution Structure Of The Third Kunitz Domain Of Tissue Factor Pathway Inhibitor
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2002-02-06 Classification: PROTEIN BINDING |

