Search Count: 703
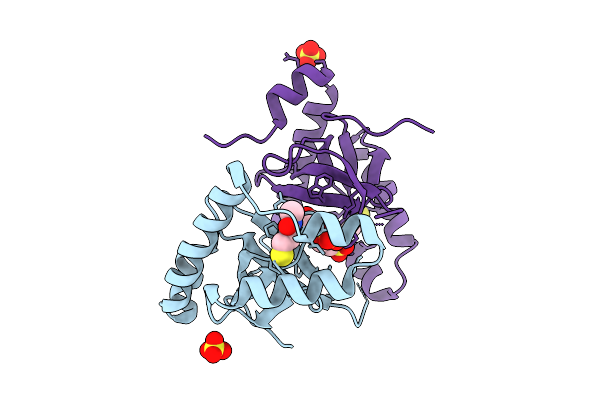 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SO4, A1EF6, EPE |
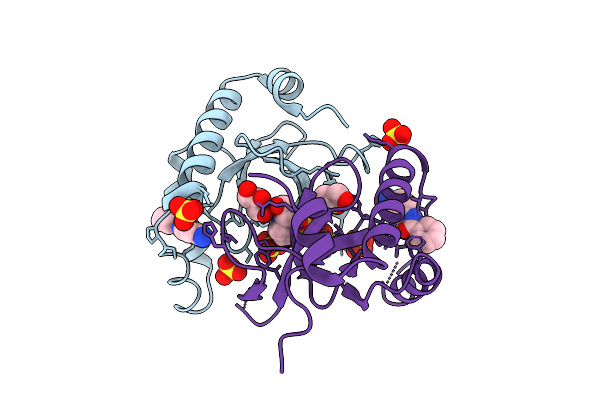 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: 6Y3, SO4, GOL, EPE |
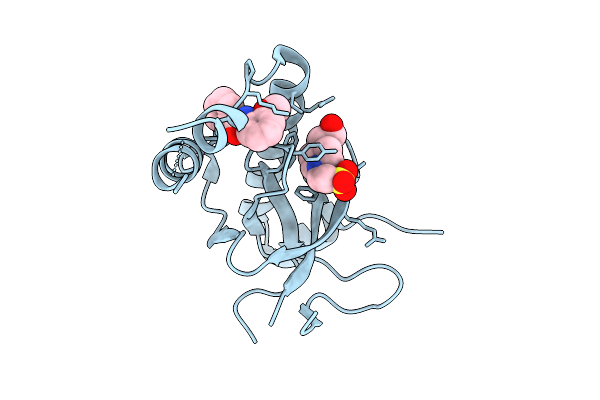 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: EPE, A1EF7 |
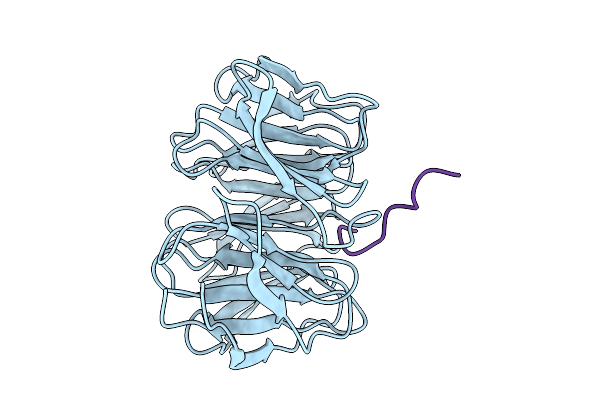 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: NUCLEAR PROTEIN |
 |
Crystal Structure Of Transaldolase Aprg From Streptoalloteichus Tenebrarius
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: PEG |
 |
Complex Crystal Structure Of Transaldolase Aprg With Glcnac From Streptoalloteichus Tenebrarius
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: A1EJ3, A1EJ2, PEG, NAG, A1EJ0 |
 |
Complex Crystal Structure Of Transaldolase Aprg With Galnac From Streptoalloteichus Tenebrarius
Organism: Streptoalloteichus tenebrarius
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: PEG |
 |
Organism: Parasutterella secunda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Parasutterella secunda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GDP, PEG |
 |
Organism: Parasutterella secunda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GNP |
 |
Crystal Structure Of Pichia Pastoris-Expressed Fast-Petase-N212A/K233C/S282C Variant
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: NAG |

