Search Count: 15
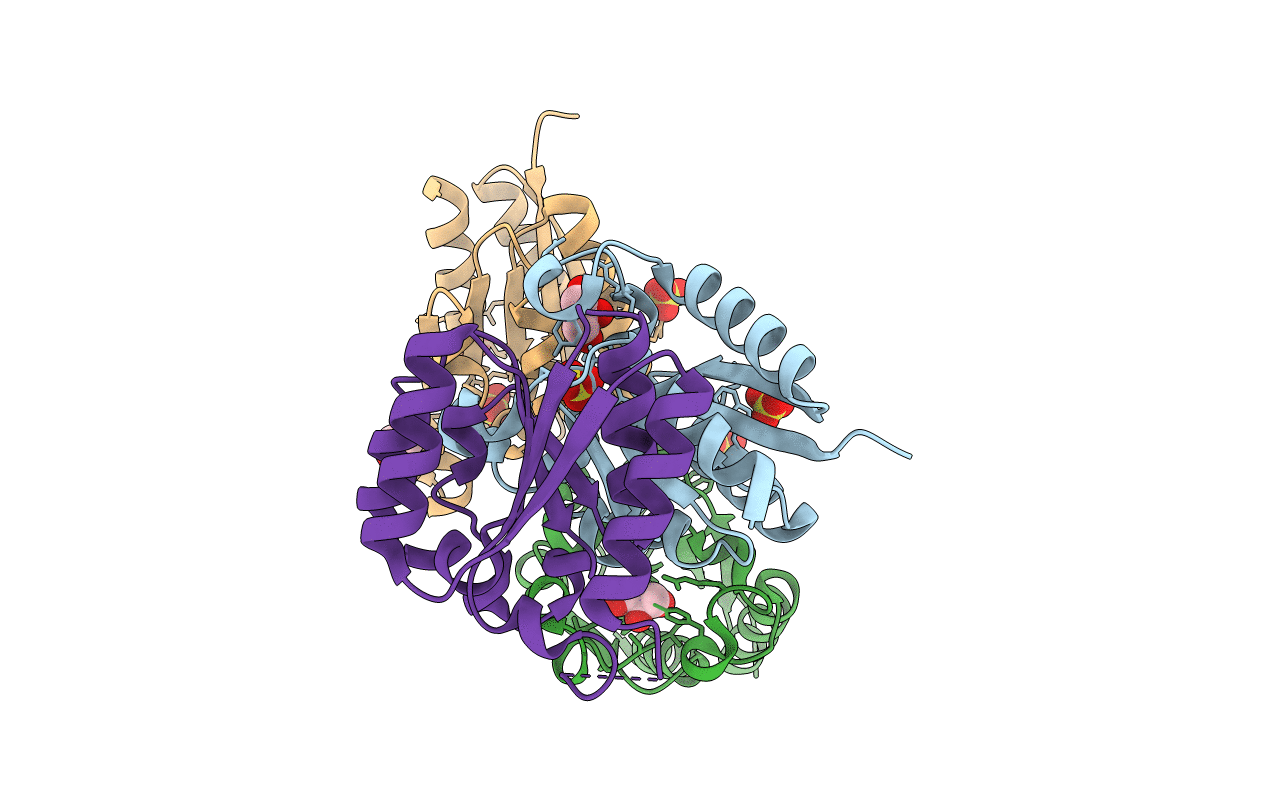 |
The Crystal Structure Of Type Ii Dehydroquinase From Propionibacterium Acnes
Organism: Cutibacterium acnes
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-07-08 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL |
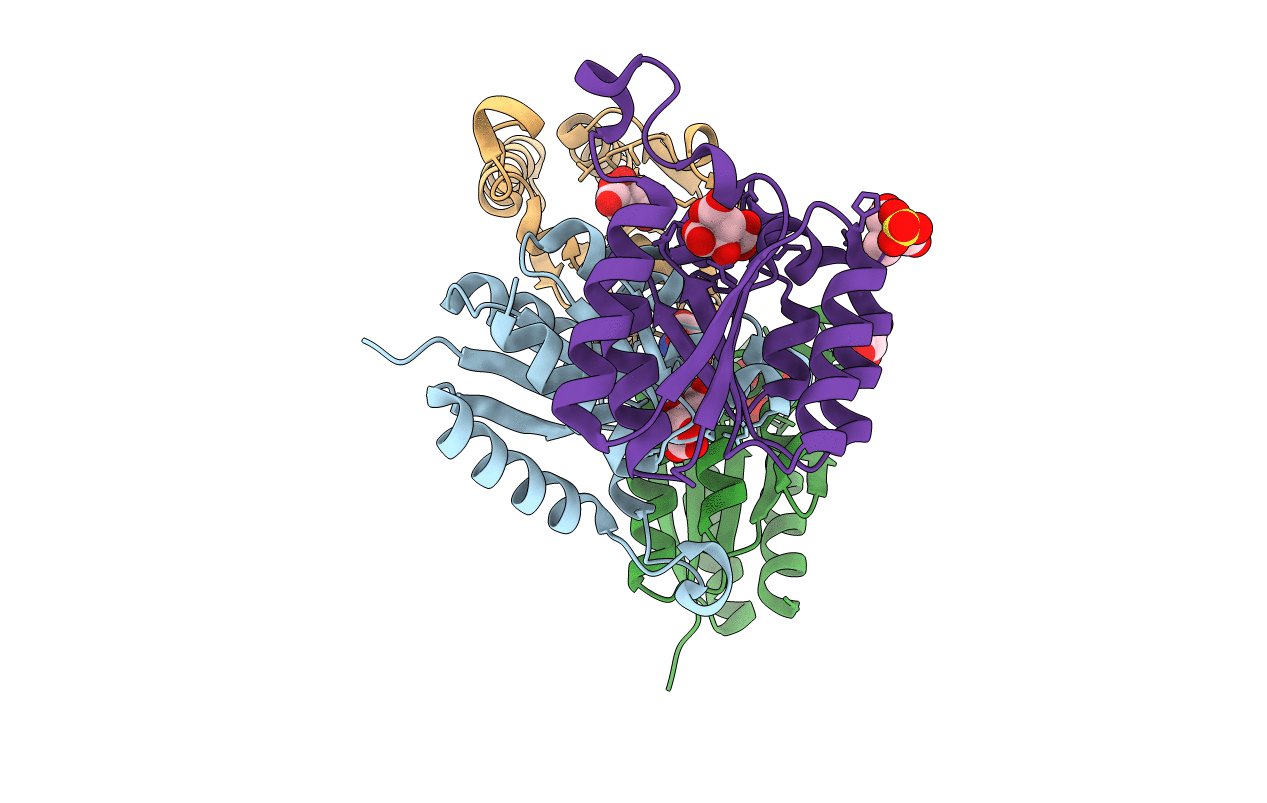 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-07-08 Classification: BIOSYNTHETIC PROTEIN Ligands: FLC, TRS, SO4, GOL |
 |
The Crystal Structure Of The Ferredoxin Receptor Fusa From Pectobacterium Atrosepticum Scri1043
Organism: Pectobacterium atrosepticum
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-08-31 Classification: TRANSPORT PROTEIN Ligands: LDA, BOG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2016-08-31 Classification: ELECTRON TRANSPORT Ligands: FES, CL |
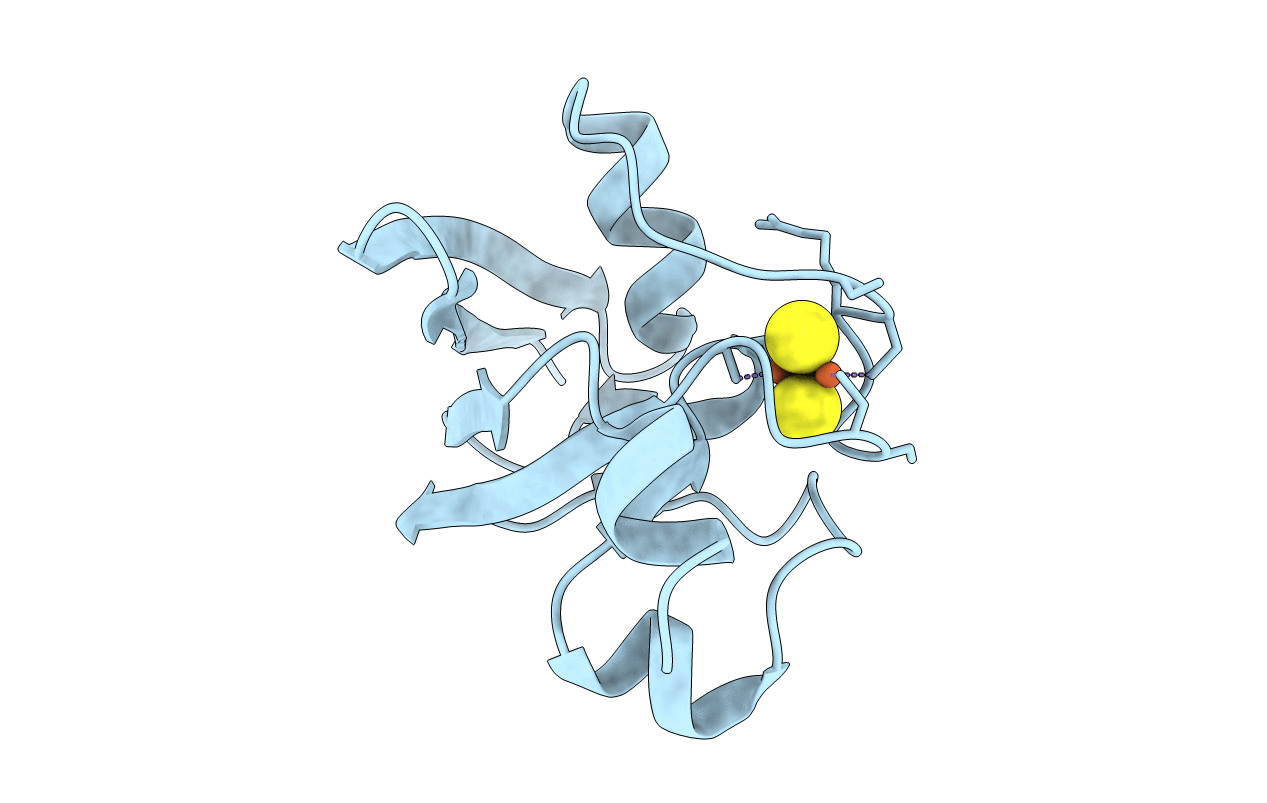 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-08-31 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Organism: Pectobacterium carotovorum subsp. brasiliensis pbr1692
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2014-06-04 Classification: HYDROLASE Ligands: FES, SO4, GOL, MPD, CL |
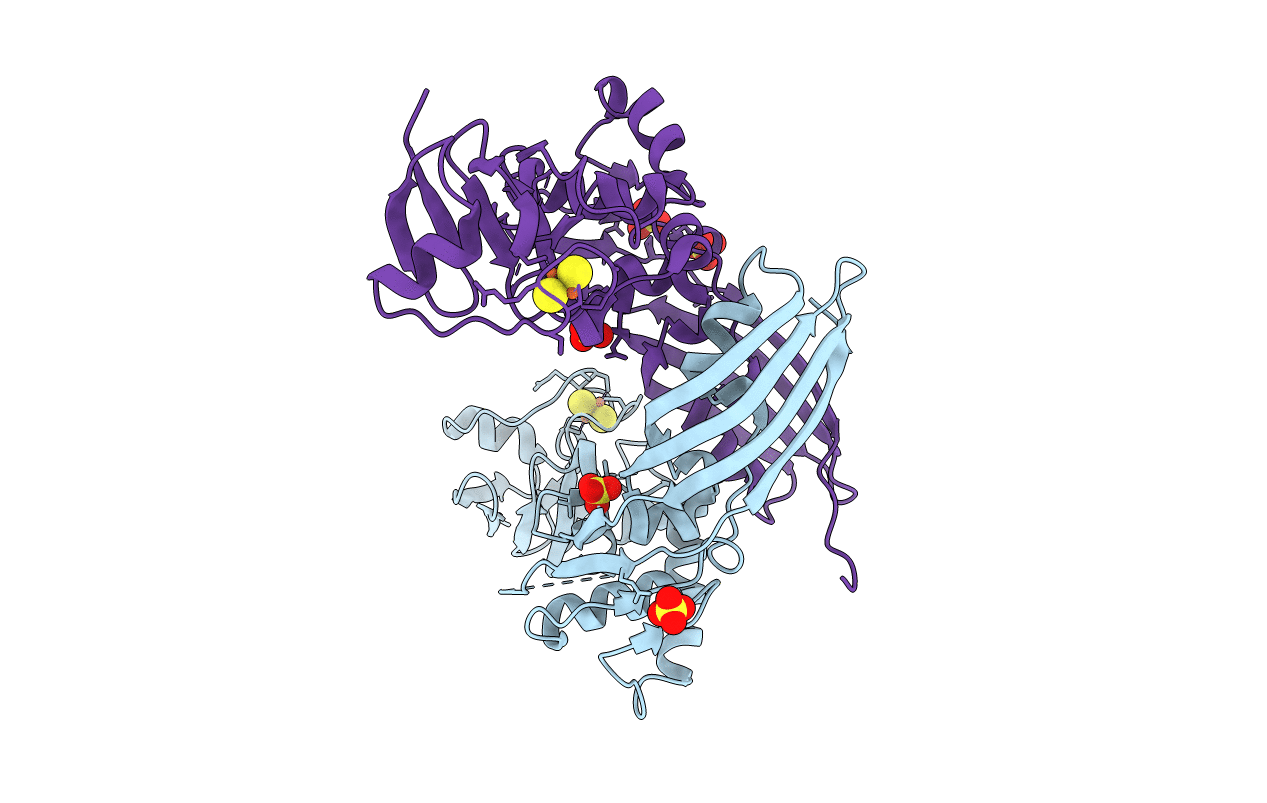 |
Organism: Pectobacterium carotovorum subsp. brasiliensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-06-04 Classification: HYDROLASE Ligands: FES, SO4, CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2014-02-19 Classification: SUGAR BINDING PROTEIN Ligands: EDO, CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2014-02-19 Classification: SUGAR BINDING PROTEIN Ligands: BMA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2014-02-19 Classification: SUGAR BINDING PROTEIN Ligands: XXR |
 |
High Resolution Structure Of Truncated Bacteriocin Syringacin M From Pseudomonas Syringae Pv. Tomato Dc3000
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2012-10-03 Classification: ANTIMICROBIAL PROTEIN Ligands: CA, EDO, CL, MG |
 |
Crystal Structure Of The Bacteriocin Syringacin M From Pseudomonas Syringae Pv. Tomato Dc3000
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2012-10-03 Classification: ANTIMICROBIAL PROTEIN Ligands: CA, EDO |
 |
Crystal Structure Of Syringacin M Mutant D232A From Pseudomonas Syringae Pv. Tomato Dc3000
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2012-10-03 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-10-11 Classification: REPLICATION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-10-04 Classification: REPLICATION |

