Search Count: 251
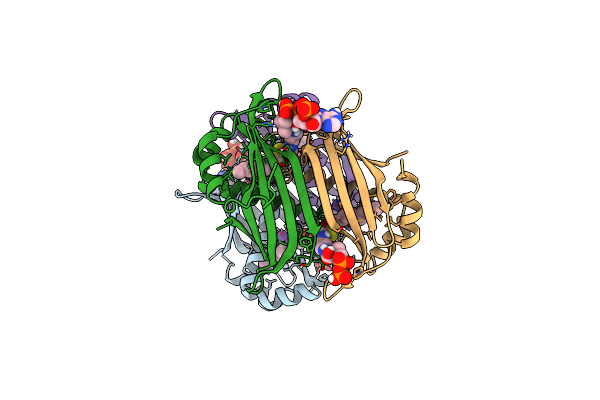 |
Crystal Structure Of The Espe7 Thioesterase Mutant R35A From The Esperamicin Biosynthetic Pathway At 1.6 A
Organism: Actinomadura verrucosospora
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-06-25 Classification: HYDROLASE Ligands: K, DCC |
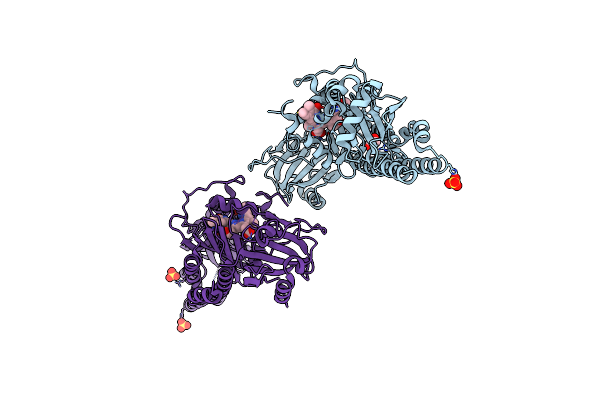 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
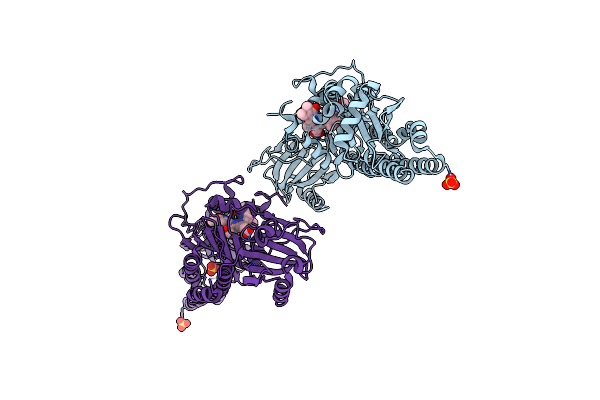 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
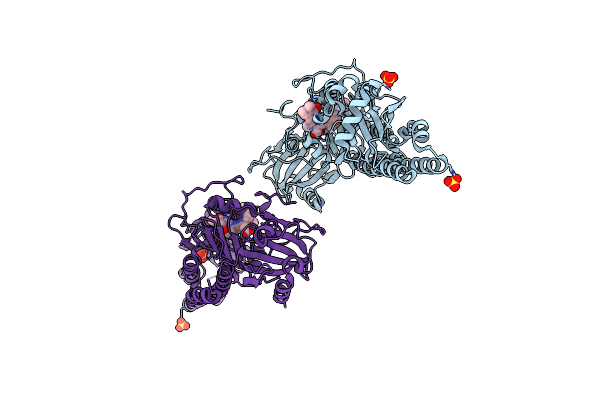 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
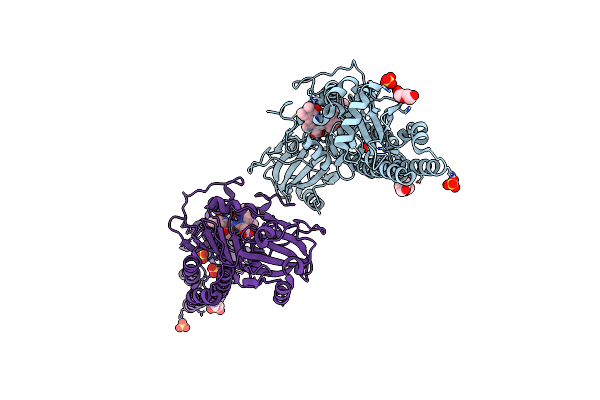 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
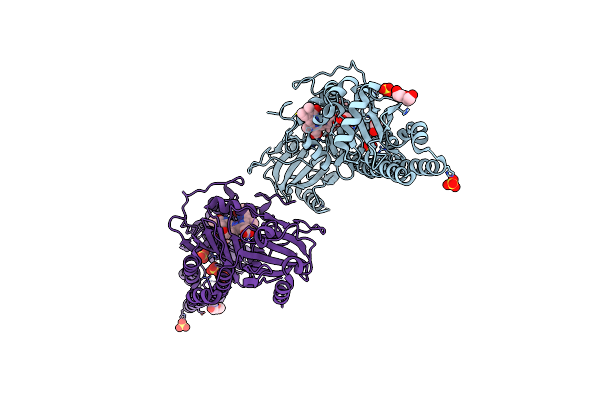 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
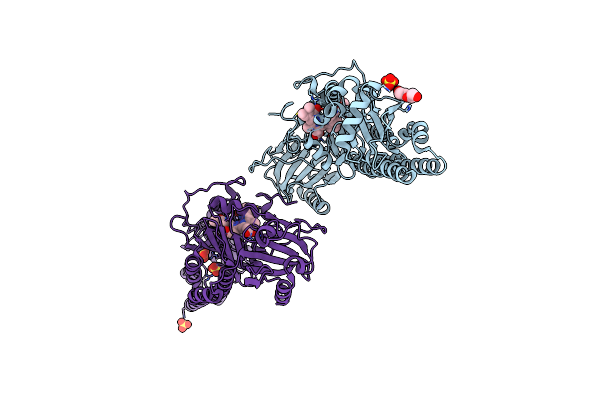 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
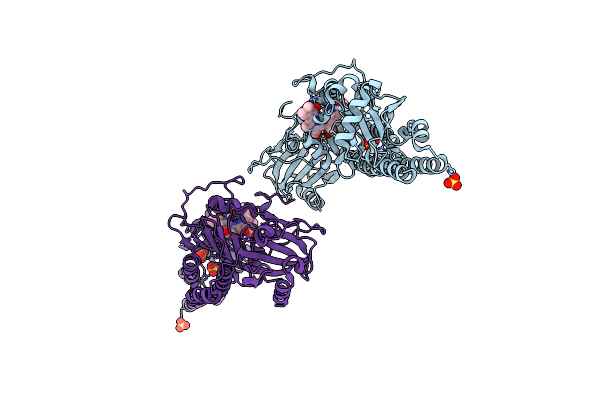 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
 |
Crystal Structure Of A Synthetic Abc Heterotrimeric Collagen-Like Peptide At 1.53 A
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2024-05-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Organism: Synechococcus sp. ja-2-3b'a(2-13)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-05-22 Classification: GENE REGULATION Ligands: CYC, EDO, ACT |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION Ligands: GOL, EDO |
 |
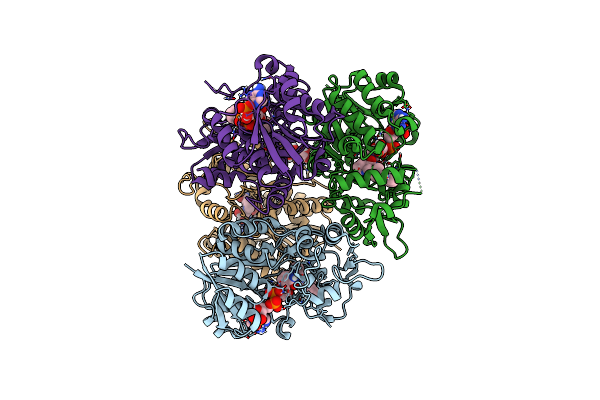
Crystal Structure Of The N-Terminal Domain Of The Cryptic Surface Protein (Cd630_25440) From Clostridium Difficile.
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-10 Classification: UNKNOWN FUNCTION Ligands: CL, EDO, GOL |
 |
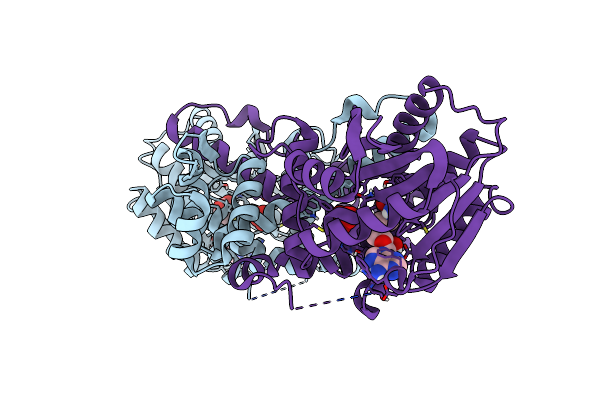
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: NAP, N3L, EDO |
 |
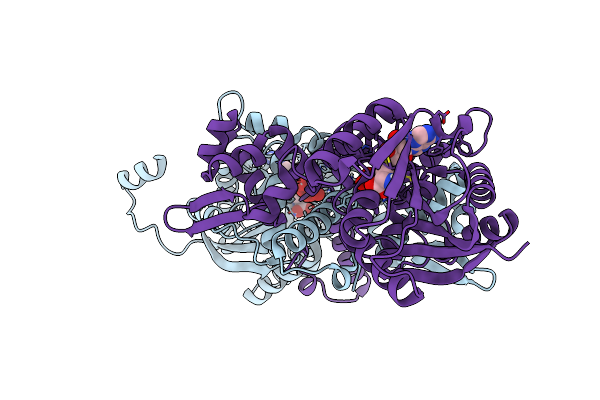
Crystal Structure Of Mfng, An L- And D-Tyrosine O-Methyltransferase From The Marformycin Biosynthesis Pathway Of Streptomyces Drozdowiczii, With Sah Bound At 1.35 A Resolution (P212121 - Form I)
Organism: Streptomyces drozdowiczii
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-10-12 Classification: TRANSFERASE Ligands: SAH, UNL |
 |
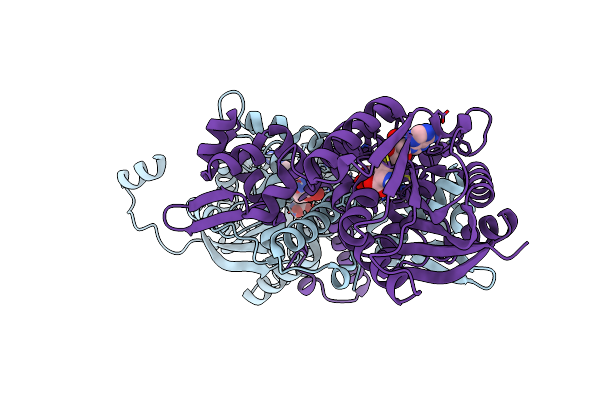
Crystal Structure Of Mfng, An L- And D-Tyrosine O-Methyltransferase From The Marformycin Biosynthesis Pathway Of Streptomyces Drozdowiczii, With Sah Bound At 1.2 A Resolution (P212121 - Form Ii)
Organism: Streptomyces drozdowiczii
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-10-12 Classification: TRANSFERASE Ligands: SAH, UNL |
 |
Crystal Structure Of Mfng, An L- And D-Tyrosine O-Methyltransferase From The Marformycin Biosynthesis Pathway Of Streptomyces Drozdowiczii, With Sah And L-Tyrosine Bound At 1.4 A Resolution (P212121 - Form Ii)
Organism: Streptomyces drozdowiczii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: SAH, TYR, UNL |
 |
Crystal Structure Of Ttnm, A Fe(Ii)-Alpha-Ketoglutarate-Dependent Hydroxylase From The Tautomycetin Biosynthesis Pathway In Streptomyces Griseochromogenes At 2 A.
Organism: Streptomyces griseochromogenes
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-07-06 Classification: OXIDOREDUCTASE Ligands: FE2, CL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-22 Classification: STRUCTURAL PROTEIN Ligands: PO4, 9F2 |

