Search Count: 26
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-03-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: U9Y |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-10-16 Classification: DNA BINDING PROTEIN INHIBITOR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-09-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-18 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of O-Acetylserine Sulfhydrylase (Tm0665) From Thermotoga Maritima At 1.80 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-09-02 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Alcohol Dehydrogenase, Iron-Containing (Tm0920) From Thermotoga Maritima At 1.30 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2003-06-10 Classification: OXIDOREDUCTASE Ligands: FE, NAP, TRS |
 |
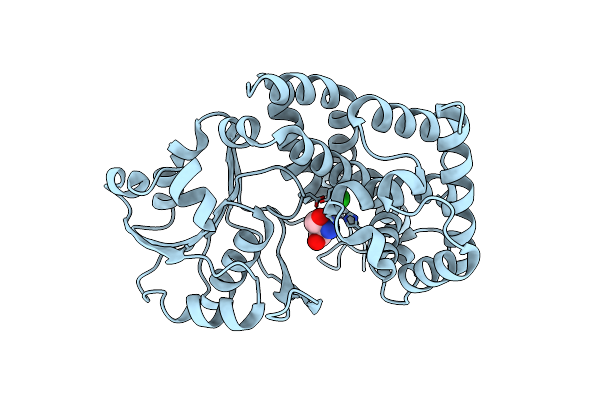
Crystal Structure Of Uronate Isomerase (Tm0064) From Thermotoga Maritima At 2.85 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2002-07-17 Classification: ISOMERASE |
 |
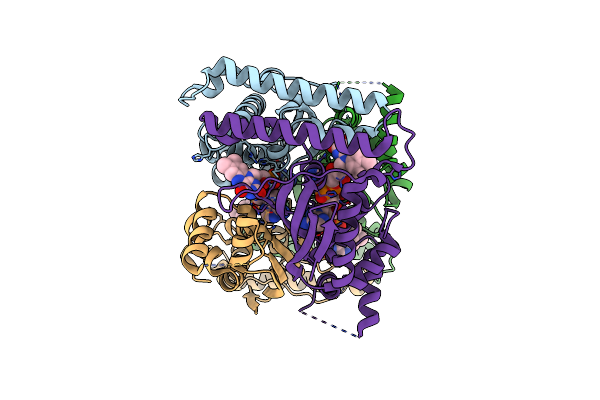
Crystal Structure Of A Glycerol Dehydrogenase (Tm0423) From Thermotoga Maritima At 1.5 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-02-27 Classification: OXIDOREDUCTASE Ligands: ZN, CL, TRS |
 |
Crystal Structure Of A Thy1-Complementing Protein (Tm0449) From Thermotoga Maritima At 2.25 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2002-02-27 Classification: TRANSFERASE Ligands: FAD |
 |
Effect Of Unnatural Heme Substitution On Kinetics Of Electron Transfer In Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-01-13 Classification: OXIDOREDUCTASE Ligands: DDH |
 |
Interaction Between Proximal And Distals Regions Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1998-10-21 Classification: PEROXIDASE Ligands: HEM |
 |
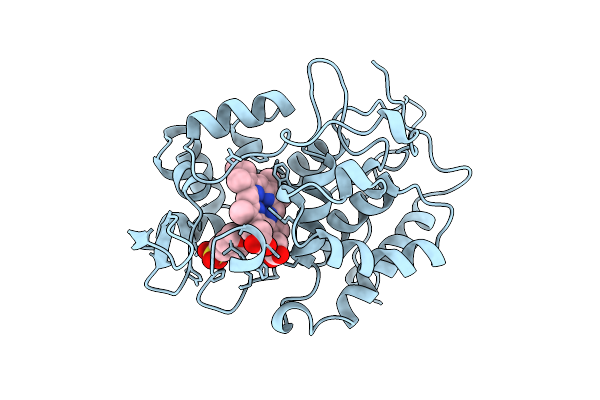
Effect Of Unnatural Heme Substitution On Kinetics Of Electron Transfer In Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1998-10-21 Classification: PEROXIDASE Ligands: HEM |
 |
Interaction Between Proximal And Distals Regions Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1998-10-21 Classification: PEROXIDASE Ligands: HEM, MES |
 |
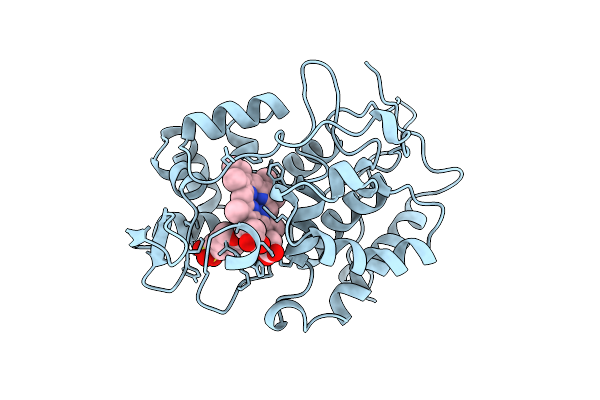
Effect Of Unnatural Heme Substitution On Kinetics Of Electron Transfer In Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1998-10-21 Classification: PEROXIDASE Ligands: CCH |
 |
Interaction Between Proximal And Distals Regions Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 1998-10-21 Classification: PEROXIDASE Ligands: HEM, MES |
 |
Interaction Between Proximal And Distals Regions Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-10-21 Classification: PEROXIDASE Ligands: CL, HEM, MES |
 |
Identifying The Physiological Electron Transfer Site Of Cytochrome C Peroxidase By Structure-Based Engineering
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 1995-12-07 Classification: OXIDOREDUCTASE (H2O2(A)) Ligands: HEM |
 |
A Cation Binding Motif Stabilizes The Compound I Radical Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-11-01 Classification: OXIDOREDUCTASE(H2O2(A)) Ligands: NH4, HEM |
 |
A Cation Binding Motif Stabilizes The Compound I Radical Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-11-01 Classification: OXIDOREDUCTASE(H2O2(A)) Ligands: K, HEM |
 |
A Cation Binding Motif Stabilizes The Compound I Radical Of Cytochrome C Peroxidase
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-11-01 Classification: OXIDOREDUCTASE(H2O2(A)) Ligands: HEM, TRS |

