Search Count: 26
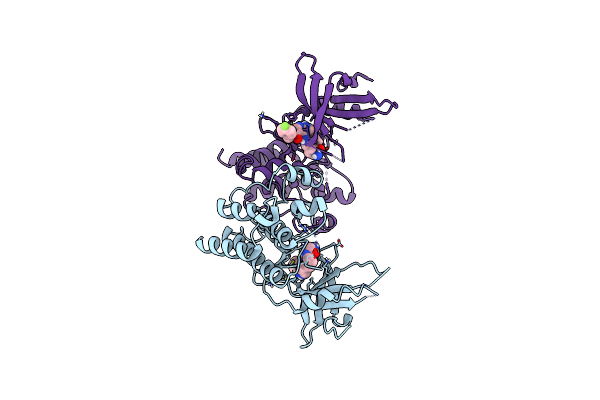 |
Crystal Structure Of The Catalytic Domain Of The Proto-Oncogene Tyrosine-Protein Kinase Mer In Complex With Inhibitor Unc2541
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2017-02-22 Classification: Transferase/Transferase Inhibitor Ligands: CL, K0X |
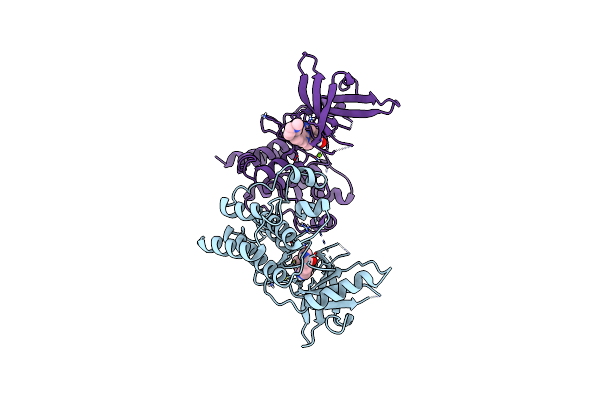 |
Crystal Structure Of The Catalytic Domain Of The Proto-Oncogene Tyrosine-Protein Kinase Mer In Complex With Inhibitor Unc2434
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-01-11 Classification: Transferase/Transferase Inhibitor Ligands: CL, 6Q1, MG |
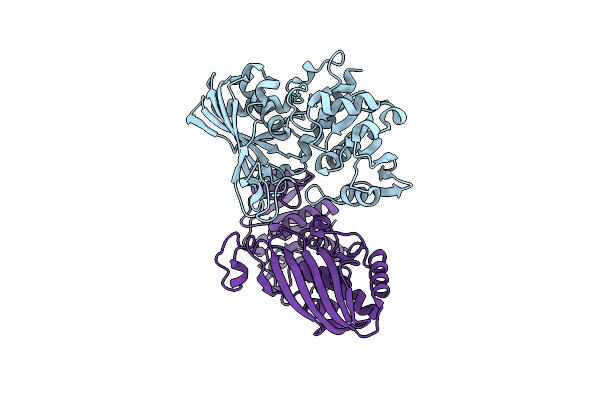 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE |
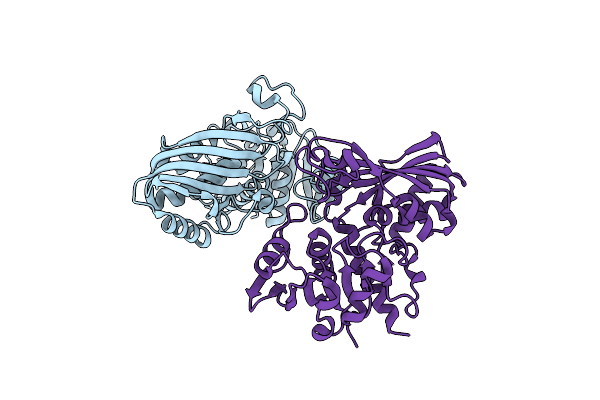 |
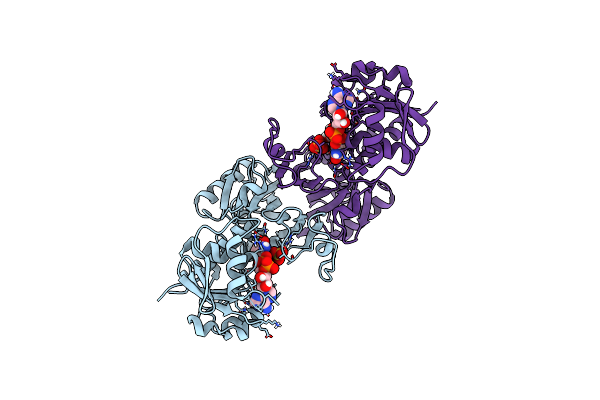
Structure Of Human Testis-Specific Glyceraldehyde-3-Phosphate Dehydrogenase Apo Form
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE |
 |
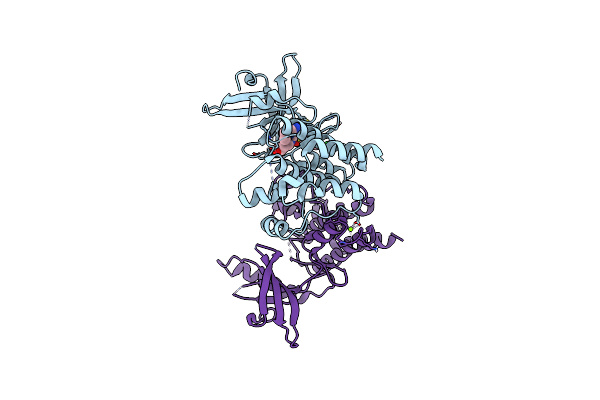
Structure Of Human Testis-Specific Glyceraldehyde-3-Phosphate Dehydrogenase Holo Form With Nad+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2016-03-09 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
 |
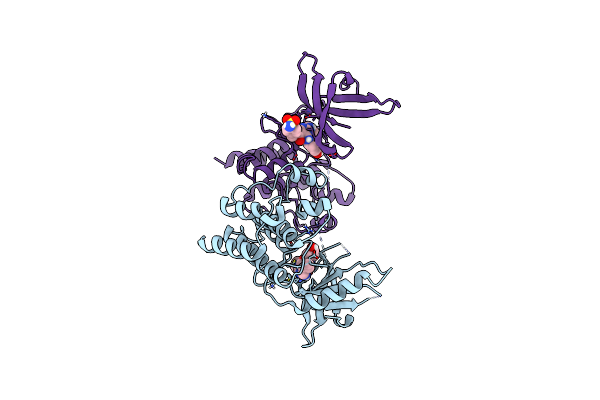
Crystal Structure Of The Catalytic Domain Of The Proto-Oncogene Tyrosine-Protein Kinase Mer In Complex With Inhibitor Unc1896
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2014-05-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: CL, MG, MH7 |
 |
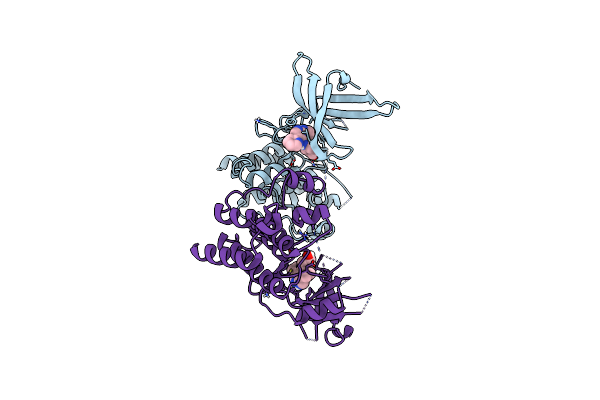
Crystal Structure Of The Catalytic Domain Of The Proto-Oncogene Tyrosine-Protein Kinase Mer In Complex With Inhibitor Unc1817
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-05-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: CL, 4MH, MG |
 |
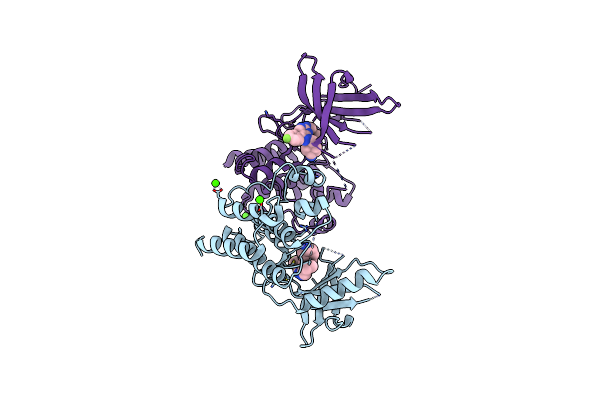
Crystal Structure Of The Catalytic Domain Of The Proto-Oncogene Tyrosine-Protein Kinase Mer In Complex With Inhibitor Unc1917
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2013-11-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: CL, MG, 24K |
 |
Crystal Structure Of The Catalytic Domain Of The Proto-Oncogene Tyrosine-Protein Kinase Mer In Complex With Inhibitor Unc569
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2012-06-20 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: CKJ, CL, CA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-06-13 Classification: UNKNOWN FUNCTION |
 |
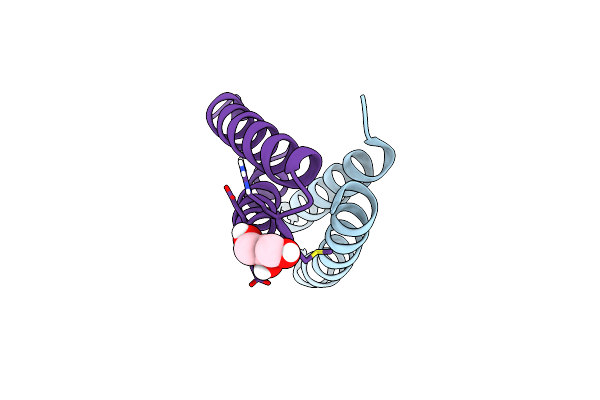
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2012-01-11 Classification: DE NOVO PROTEIN, METAL BINDING PROTEIN |
 |
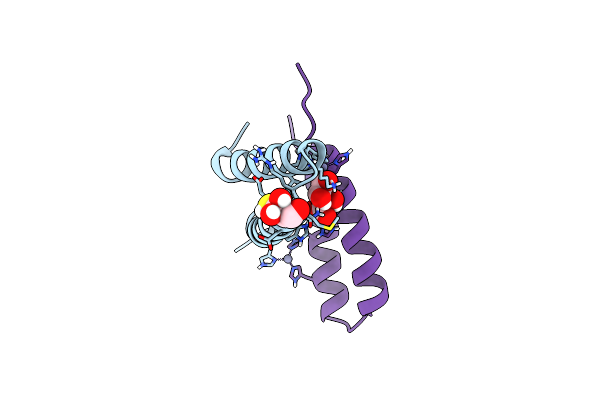
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2012-01-11 Classification: DE NOVO PROTEIN, METAL BINDING PROTEIN Ligands: GOL |
 |
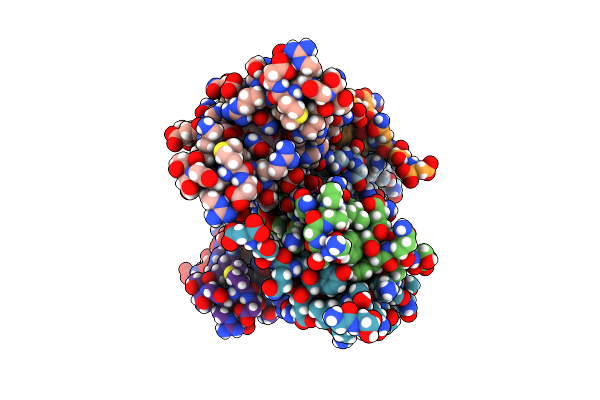
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2012-01-11 Classification: DE NOVO PROTEIN, HYDROLASE Ligands: ZN, TLA, UNL |
 |
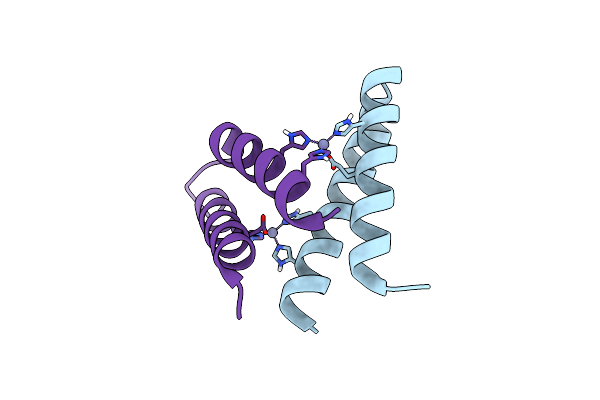
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2012-01-11 Classification: DE NOVO PROTEIN, METAL BINDING PROTEIN Ligands: CO, 1PE |
 |
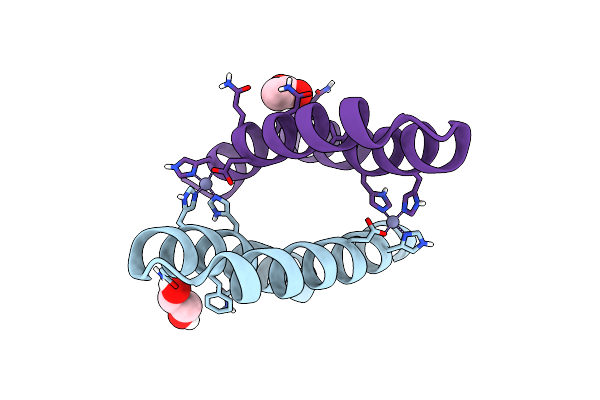
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2012-01-11 Classification: DE NOVO PROTEIN, METAL BINDING PROTEIN Ligands: ZN |
 |
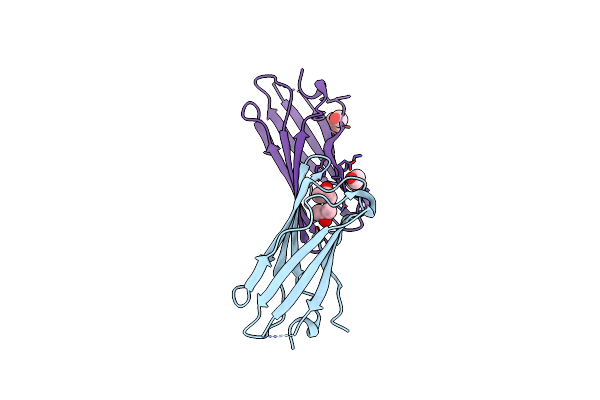
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2012-01-11 Classification: DE NOVO PROTEIN, METAL BINDING PROTEIN Ligands: ZN, EDO, ACT |
 |
Crystal Structure Of Computationally Redesigned Gamma-Adaptin Appendage Domain Forming A Symmetric Homodimer
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2011-12-28 Classification: ENDOCYTOSIS Ligands: IPA, PEG |
 |
Structure Of A G-Alpha-I1 Mutant With Enhanced Affinity For The Rgs14 Goloco Motif.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2010-11-24 Classification: SIGNALING PROTEIN Ligands: GDP, SO4 |
 |
Crystal Structure Of The Udp-Glucuronic Acid Binding Domain Of The Human Drug Metabolizing Udp-Glucuronosyltransferase 2B7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-05-01 Classification: TRANSFERASE |
 |
Mhc Class I Natural Mutant H-2Kbm8 Heavy Chain Complexed With Beta-2 Microglobulin And Herpes Simplex Virus Glycoprotein B Peptide
Organism: Mus musculus, Herpes simplex virus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-12-14 Classification: IMMUNE SYSTEM |

