Search Count: 37
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, FME, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
Bacteriorhodopsin Ground State Structure Obtained With Serial Femtosecond Crystallography
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-31 Classification: PROTON TRANSPORT Ligands: RET, LI1 |
 |
Serial Time Resolved Crystallography Of Photosystem Ii Using A Femtosecond X-Ray Laser. The S State After Two Flashes (S3)
Organism: Thermosynechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2015-11-04 Classification: OXIDOREDUCTASE Ligands: OEX, FE2, CLA, PHO, BCR, SQD, CL, BCT, PL9, LMG, CA, DGD, LHG, HEM, MG |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
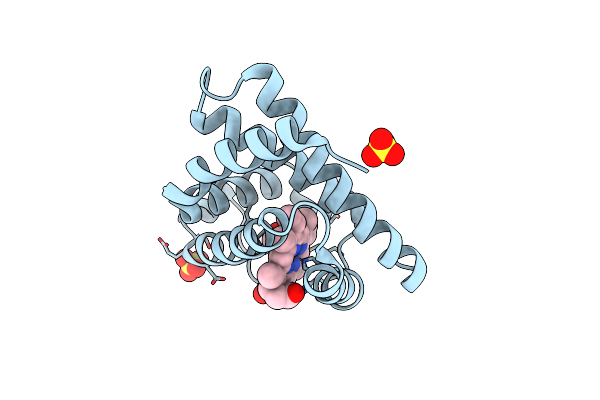
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
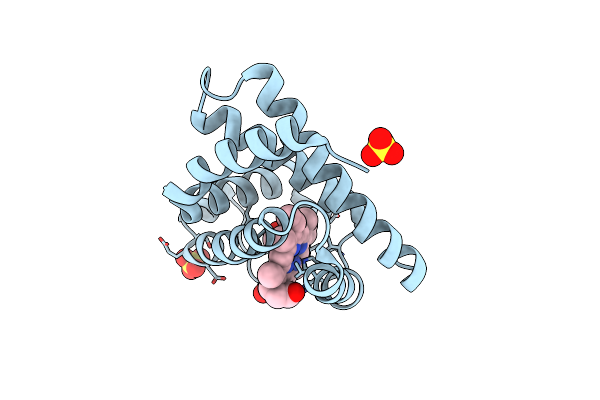
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
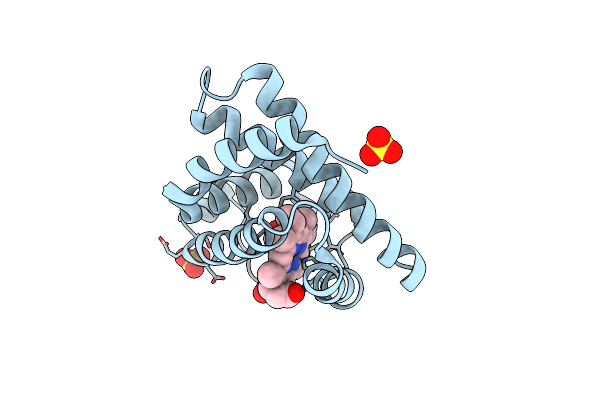
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
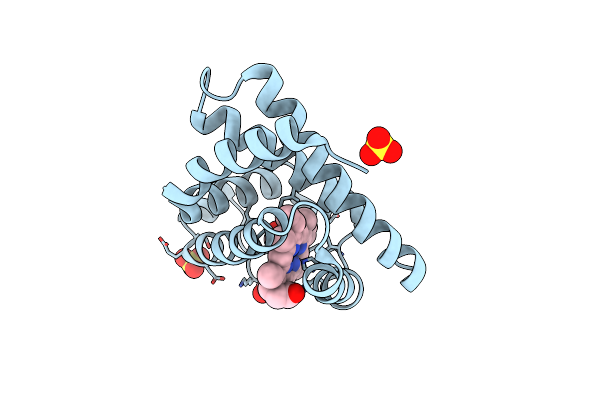
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: SO4, CMO, HEM |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-16 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CMO |

