Search Count: 25
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: 1PE, PEG, EDO, NAG, XYP |
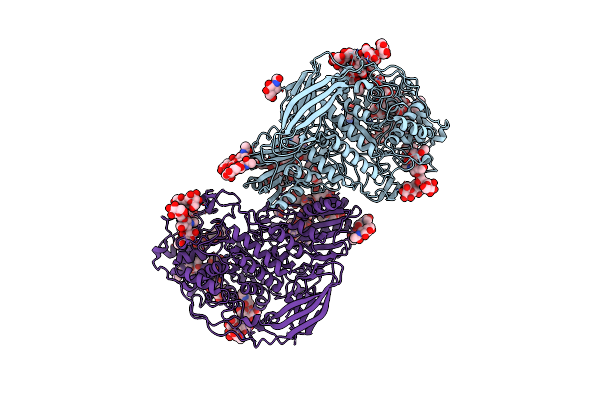 |
Structural Studies Of A Glycoside Hydrolase Family 3 Beta-Glucosidase From The Model Fungus Neurospora Crassa
Organism: Neurospora crassa or74a
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: NAG |
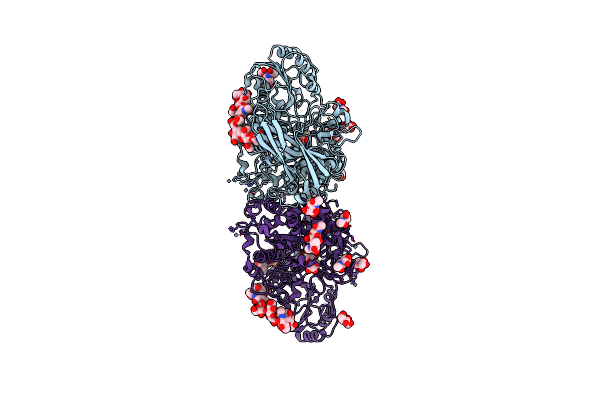 |
The Structure Of Hypocrea Jecorina Beta-Xylosidase Xyl3A (Bxl1) In Complex With 4-Thioxylobiose
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: NAG, ZN |
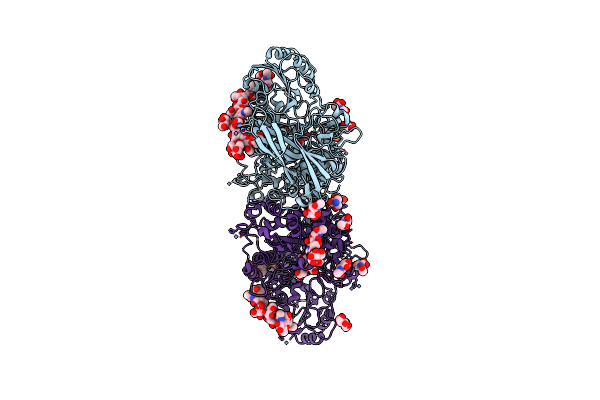 |
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-08-10 Classification: HYDROLASE Ligands: NAG, ZN, TRS, GOL, ACT |
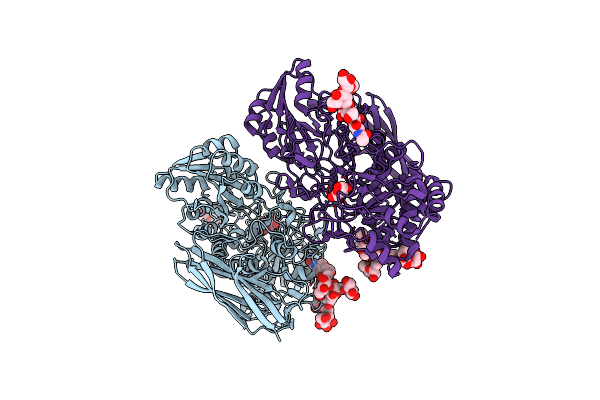 |
Crystal Structure Of Beta-D-Glucoside Glucohydrolase From Trichoderma Reesei
Organism: Trichoderma reesei
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2012-12-19 Classification: HYDROLASE Ligands: NAG, BGC |
 |
Crystal Structure Of A Glycoside Hydrolase Family 3 Beta-Glucosidase, Bgl1 From Hypocrea Jecorina At 2.1A Resolution.
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-12 Classification: HYDROLASE Ligands: NAG, BGC, GOL, PEG |
 |
Crystal Structure Of A Glycoside Hydrolase Family 3 Beta-Glucosidase, Bgl1 From Hypocrea Jecorina At 2.1A Resolution.
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-12 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Organism: Heterobasidion annosum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-04-25 Classification: HYDROLASE Ligands: MG |
 |
Elucidation Of The Substrate Specificity And Protein Structure Of Abfb, A Family 51 Alpha-L-Arabinofuranosidase From Bifidobacterium Longum.
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-12-28 Classification: HYDROLASE |
 |
Organism: Heterobasidion annosum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: EPE, XYS, MG |
 |
Drosophila Melanogaster Deoxyribonucleoside Kinase Successfully Activates Gemcitabine In Transduced Cancer Cell Lines
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-24 Classification: TRANSFERASE Ligands: GEO, SO4 |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: SO4, AZZ |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: MG, TTP |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: DGT |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: DCP |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: SO4, 5FU |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: SO4, DOC |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-04-29 Classification: TRANSFERASE Ligands: SO4, BVD |
 |
Structural Studies Of Nucleoside Analog And Feedback Inhibitor Binding To Drosophila Melanogaster Multisubstrate Deoxyribonucleoside Kinase
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-04-18 Classification: TRANSFERASE Ligands: SO4, DCZ |

