Search Count: 378
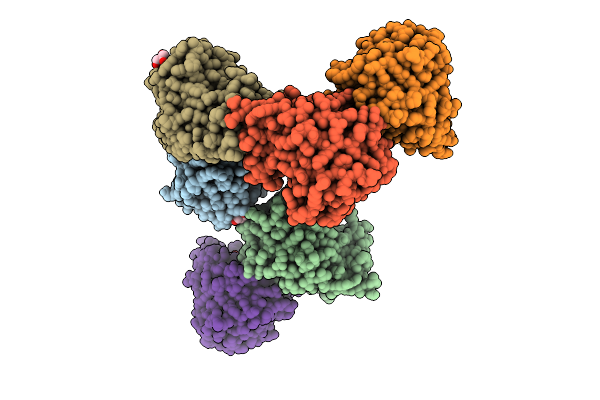 |
Crystal Structure Of Bacteroides Ovatus Kdui1 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: ISOMERASE Ligands: GOL |
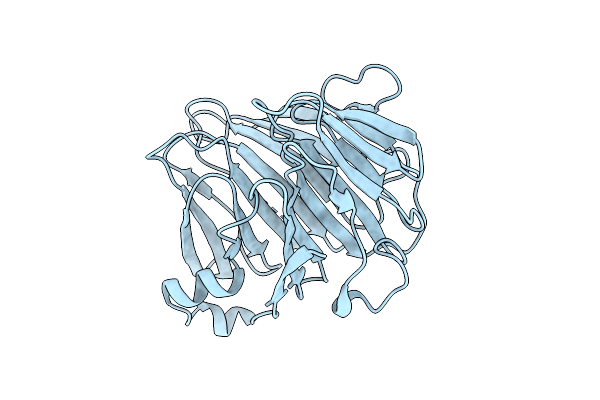 |
Crystal Structure Of Bacteroides Ovatus Kdui2 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: ISOMERASE |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Complexed With Nad+ Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: NAD |
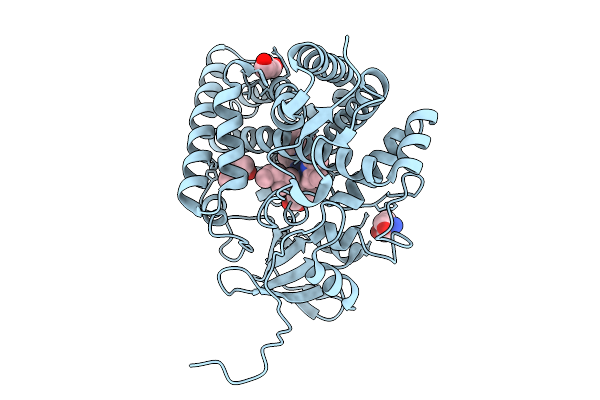 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM, MPD, TRS, PEG |
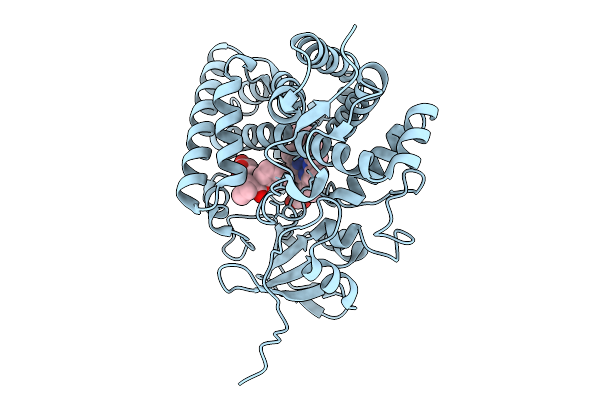 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, 114 |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, SIM, MPD |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: HEM, MPD, TRS, PEG |
 |
Crystal Structure Of The Enzyme-Product Complex Of L-Azetidine-2-Carboxylate Hydrolase
Organism: Pseudomonas sp. a2c
Method: X-RAY DIFFRACTION Resolution:0.93 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: 42B, IMD, FMT, EDO, PEG, MG |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, KTN, TRS |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, A1L6Q |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, A1L6X, TRS |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, A1L6Q |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, TRS |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: HEM, TRS, FLF, EDO |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: HEM, DIF, TRS, EDO |
 |
Organism: Streptomyces griseolus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: HEM, FLF, EDO |
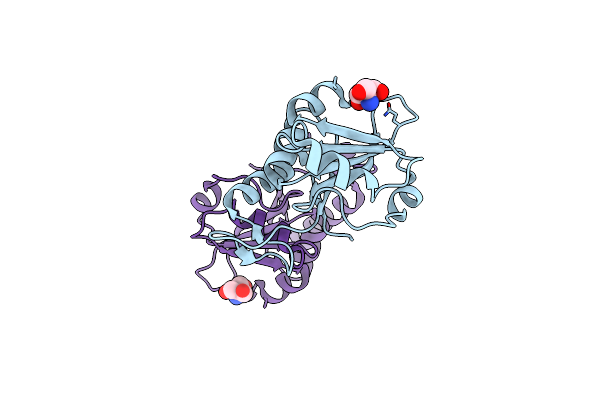 |
Crystal Structure Of Sugar Phosphotransferase System Eiib Component Cpf_0401 From Clostridium Perfringens
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: TRS |
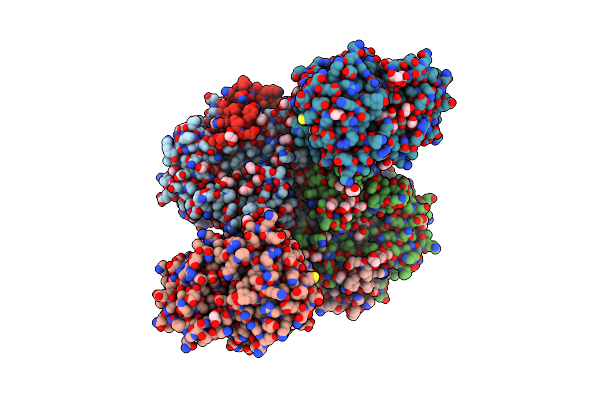 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-09 Classification: LYASE Ligands: PEG, EDO |
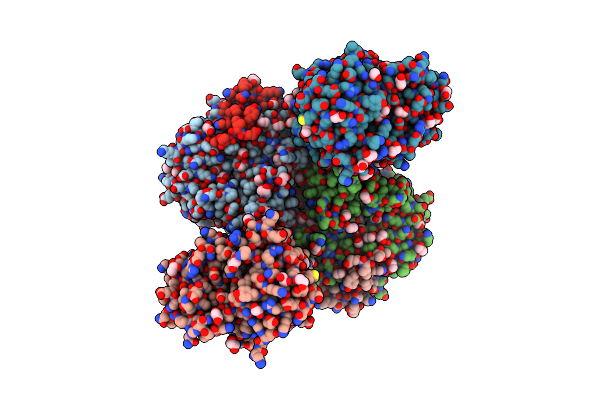 |
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-09 Classification: LYASE Ligands: PEG, EDO, AG2 |

