Search Count: 94
 |
Organism: Trypanosoma brucei brucei
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: RNA BINDING PROTEIN Ligands: 7MG |
 |
Organism: Trypanosoma brucei brucei, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: RNA BINDING PROTEIN |
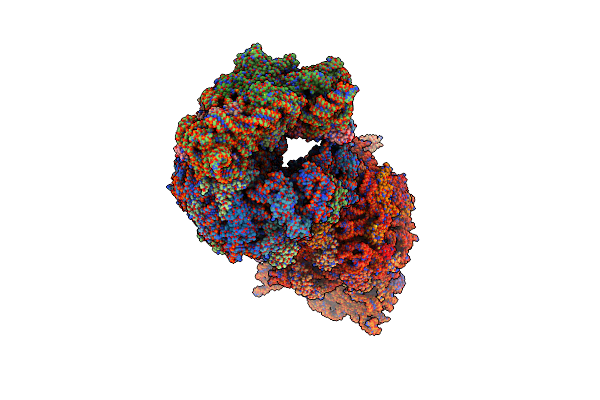 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Formyl-Mai-Tripeptidyl-Trna Analog Acca-Iamf At 2.65A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-07-19 Classification: RIBOSOME/RNA Ligands: MG, MPD, ARG, ZN, SF4 |
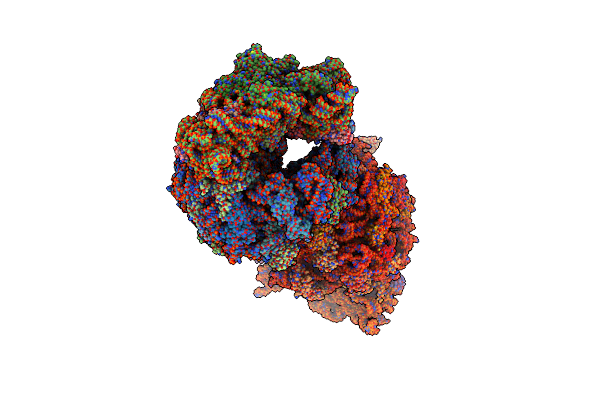 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Formyl-Mfi-Tripeptidyl-Trna Analog Acca-Ifmf At 2.60A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-19 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
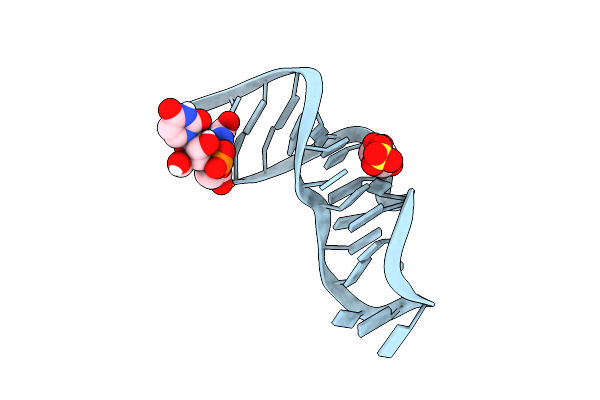 |
23S Ribosomal Rna Sarcin Ricin Loop 27-Nt Fragment Containing A Xanthosine Residue At Position 2648
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.86 Å Release Date: 2023-01-25 Classification: RNA Ligands: URI, HYJ, GOL, SO4 |
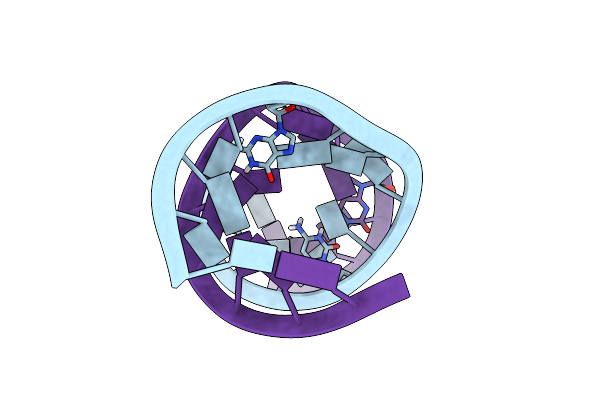 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-01-25 Classification: RNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2023-01-25 Classification: RNA |
 |
Organism: Streptococcus sp., Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-01-18 Classification: PROTEIN/RNA Ligands: NAD, MG |
 |
Organism: Streptococcus sp., Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-18 Classification: PROTEIN/RNA Ligands: NMN, MG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2022-07-20 Classification: RNA Ligands: GOL |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Mti-Tripeptidyl-Trna Analog Acca-Itm At 2.40A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, MPD, ARG, ZN, SF4, MRD |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Mai-Tripeptidyl-Trna Analog Acca-Iam At 2.45A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Aminoacyl-Trna Analog Acc-Pmn, And P-Site Mfi-Tripeptidyl-Trna Analog Acca-Ifm At 2.50A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, MPD, ARG, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Deacylated Trna Analog Cacca, P-Site Mti-Tripeptidyl-Trna Analog Acca-Itm, And Chloramphenicol At 2.50A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, CLM, ARG, MPD, ZN, SF4 |
 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Protein Y, A-Site Deacylated Trna Analog Cacca, P-Site Mai-Tripeptidyl-Trna Analog Acca-Iam, And Chloramphenicol At 2.40A Resolution
Organism: Escherichia coli k-12, Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-26 Classification: RIBOSOME Ligands: MG, CLM, ARG, MPD, ZN, SF4 |
 |
Crystal Structure Of Xanthine Riboswitch With Xanthine, Iridium Hexammine Soak
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, IR, XAN, MG |
 |
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, XAN, MN, MG |
 |
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, XAN, MG |
 |
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, MG, AZA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.01 Å Release Date: 2021-05-05 Classification: RNA Ligands: GOL |

