Search Count: 207
All
Selected
 |
Catalase Cryoem Structure From Micrococcus Luteus At 1.9 Angstrom Resolution.
Organism: Micrococcus luteus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: HEM, NDP |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-19 Classification: OXIDOREDUCTASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-08-19 Classification: OXIDOREDUCTASE |
 |
Organism: Micrococcus luteus nctc 2665
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-10-03 Classification: TRANSFERASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-04-13 Classification: HYDROLASE |
 |
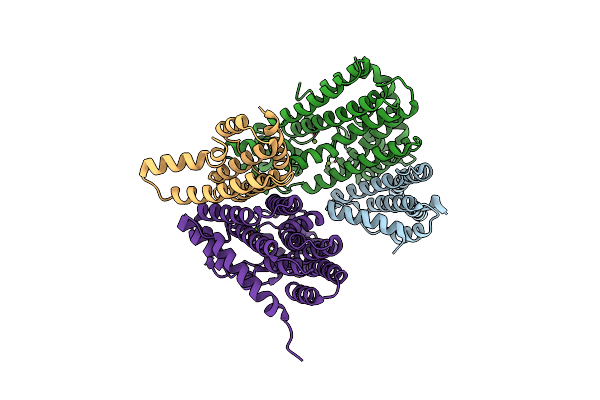
Crystal Structure Of Mglu In Its L-Glutamate Binding Form In The Presence Of 4.3M Nacl
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-04-13 Classification: HYDROLASE Ligands: GLU |
 |
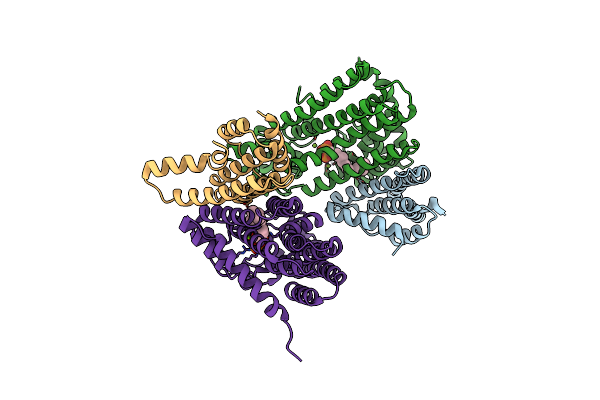
M. Luteus B-P 26 Heterodimeric Hexaprenyl Diphosphate Synthase In Complex With Magnesium
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-11-10 Classification: TRANSFERASE Ligands: MG, CL |
 |
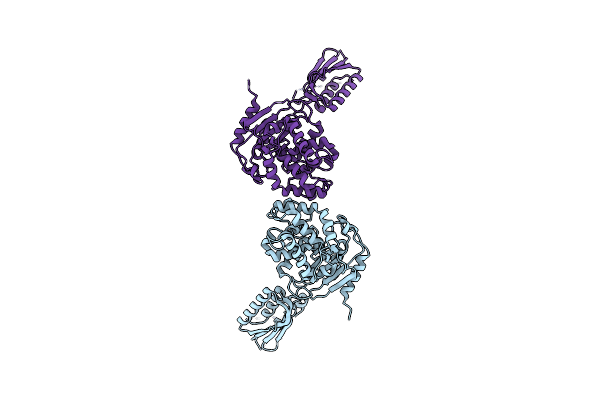
M. Luteus B-P 26 Heterodimeric Hexaprenyl Diphosphate Synthase In Complex With Magnesium And Fpp Analogue
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2010-11-10 Classification: TRANSFERASE Ligands: MG, 2DE, CL |
 |
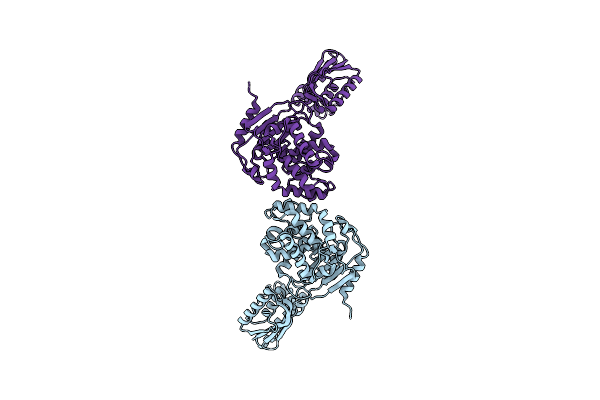
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-01-19 Classification: HYDROLASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-01-19 Classification: HYDROLASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-01-19 Classification: HYDROLASE Ligands: GLU |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-01-19 Classification: HYDROLASE Ligands: GLU, TRS |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2009-08-04 Classification: HYDROLASE |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:0.88 Å Release Date: 2002-04-26 Classification: OXIDOREDUCTASE Ligands: HEM, SO4 |
 |
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2002-04-26 Classification: OXIDOREDUCTASE Ligands: HEM, SO4, ACT, O |
 |
Atomic Resolution Structure Of Micrococcus Lysodeikticus Catalase Complexed With Nadph
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2002-03-19 Classification: OXIDOREDUCTASE Ligands: HEM, NDP, SO4, ACT |
 |
Crystal Structure Of Undecaprenyl Diphosphate Synthase From Micrococcus Luteus B-P 26
Organism: Micrococcus luteus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-03-28 Classification: TRANSFERASE Ligands: SO4 |
 |
 |
 |

