Search Count: 20
 |
Organism: Kitasatospora cystarginea
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, FLC |
 |
Crystal Structure Of Coronafacic Acid Ligase From Pectobacterium Brasiliense
Organism: Pectobacterium brasiliense
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-05-05 Classification: LIGASE Ligands: R4Z, PO4 |
 |
Crystal Structure Of The R395G Mutant Form Of Coronafacic Acid Ligase From Pectobacterium Brasiliense
Organism: Pectobacterium brasiliense
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-05-05 Classification: LIGASE Ligands: R4Z, PO4 |
 |
X-Ray Crystal Structure Of The Tetrahydrofolate Riboswitch Aptamer Bound To 5-Deazatetrahydropterin
Organism: Streptococcus mutans ua159
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2019-12-18 Classification: RNA Ligands: T0A, MG |
 |
Crystal Structure Of Coclaurine N-Methyltransferase (Cnmt) Bound To N-Methylheliamine And Sah
Organism: Coptis japonica
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of Coclaurine N-Methyltransferase (Cnmt) Bound To N-Methylheliamine And Sah
Organism: Coptis japonica
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: SAH, F2W |
 |
Crystal Structure Of Coclaurine N-Methyltransferase (Cnmt) Bound To N-Methylheliamine And Sah
Organism: Coptis japonica
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2018-06-06 Classification: TRANSFERASE Ligands: SAH, F2Z |
 |
Crystal Structure Of Thermophilic Tryptophan Halogenase (Th-Hal) Enzyme From Streptomycin Violaceusniger.
Organism: Streptomyces violaceusniger
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2016-10-19 Classification: HYDROLASE |
 |
Crystal Structure Of Thermophilic Tryptophan Halogenase (Th-Hal) Enzyme From Streptomycin Violaceusniger.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2016-10-19 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-04-13 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PO4, SO4 |
 |
Crystal Structure Of A Tryptophan 6-Halogenase (Stth) From Streptomyces Toxytricini
Organism: Streptomyces toxytricini
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2016-02-17 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
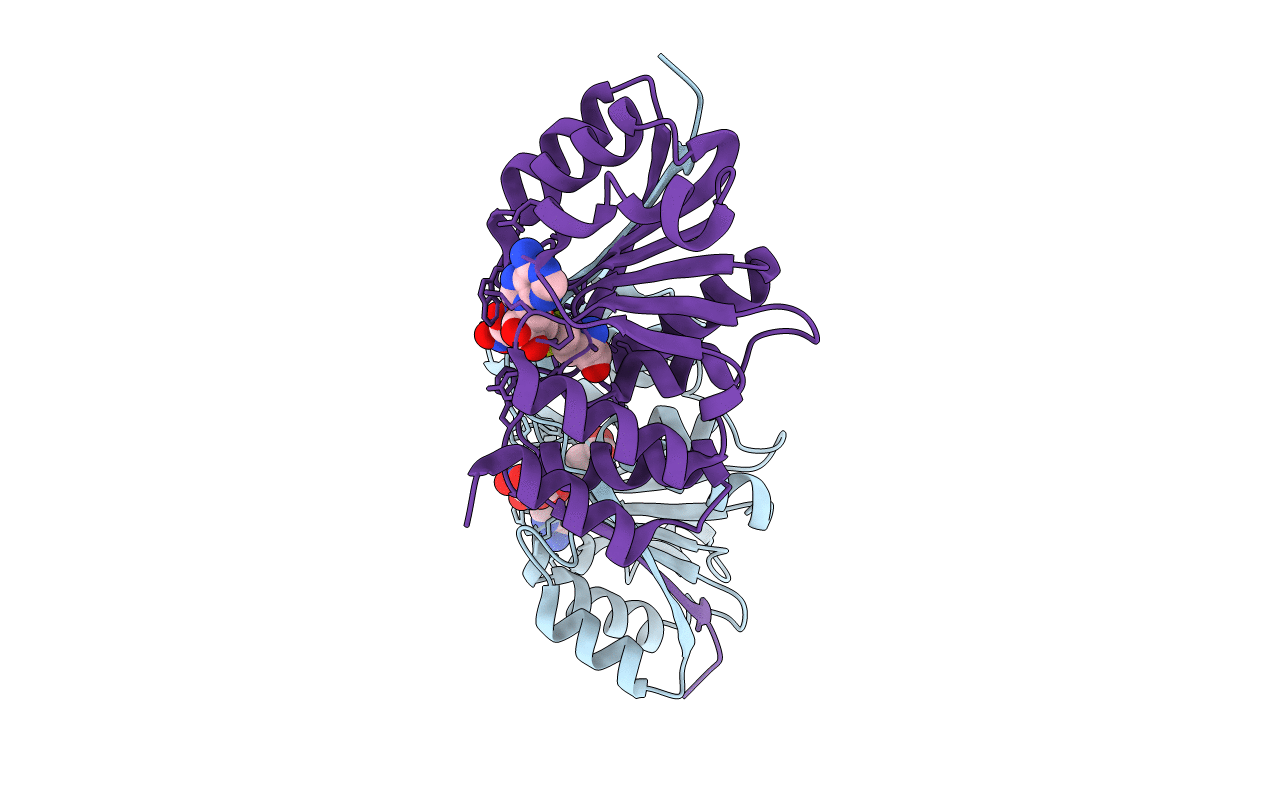
Crystal Structure Of (Wt) Rat Catechol-O-Methyltransferase In Complex With Adomet And 3,5-Dinitrocatechol (Dnc)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: SAM, DNC, MG |
 |
Crystal Structure Of Y200L Mutant Of Rat Catechol-O-Methyltransferase In Complex With Adomet And 3,5-Dinitrocatechol
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: DNC, MG, SAM, BR |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, CL |
 |
X-Ray Crystal Structure Of The M6" Riboswitch Aptamer Bound To Pyrimido[4,5-D]Pyrimidine-2,4-Diamine (Ppda)
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2014-07-16 Classification: RNA Ligands: 29G, MG |
 |
X-Ray Crystal Structure Of The M6C" Riboswitch Aptamer Bound To 2-Aminopyrimido[4,5-D]Pyrimidin-4(3H)-One (Ppao)
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-07-16 Classification: RNA Ligands: 29H, MG |
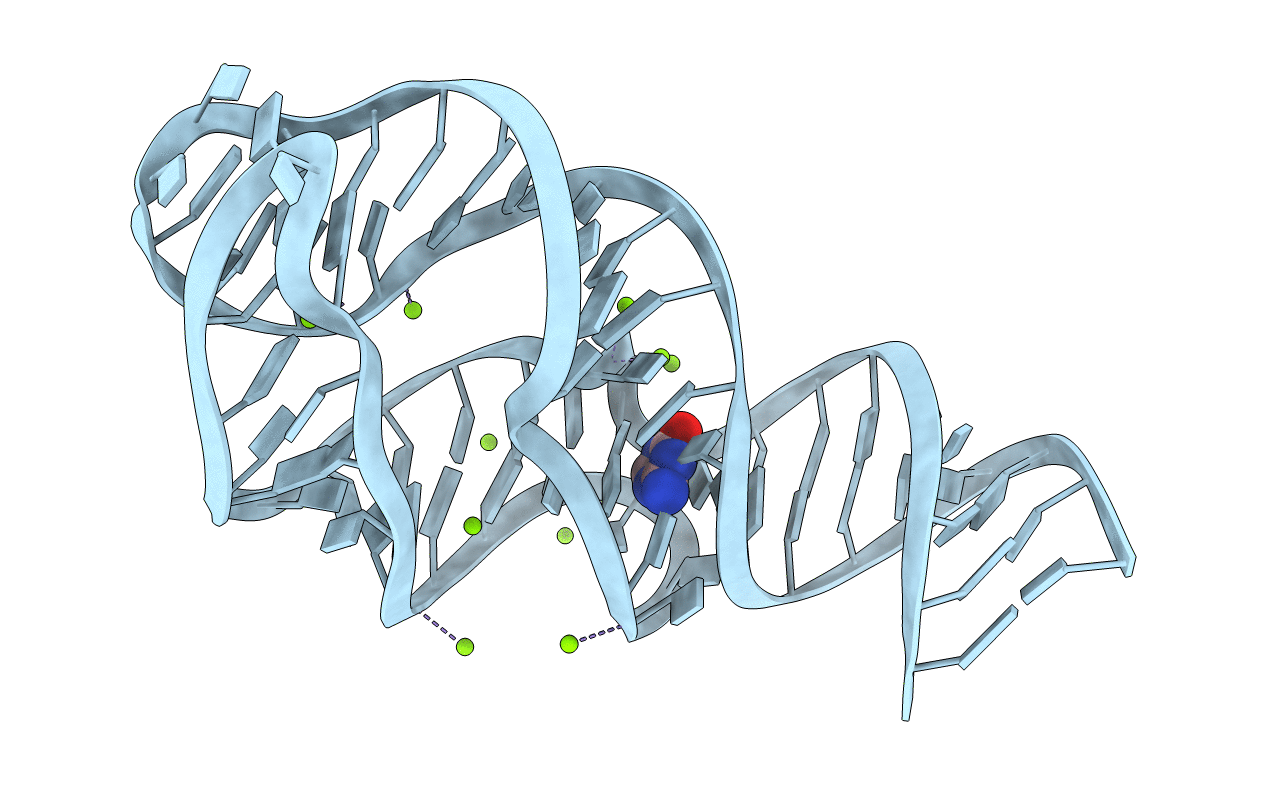 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-01-26 Classification: RNA Ligands: MG, 5AZ |
 |
Crystal Structure Of Arylmalonate Decarboxylase (Amdase) From Bordatella Bronchiseptic In Complex With Benzylphosphonate
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-10-06 Classification: LYASE Ligands: B85 |
 |
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-08-05 Classification: LYASE Ligands: PO4 |
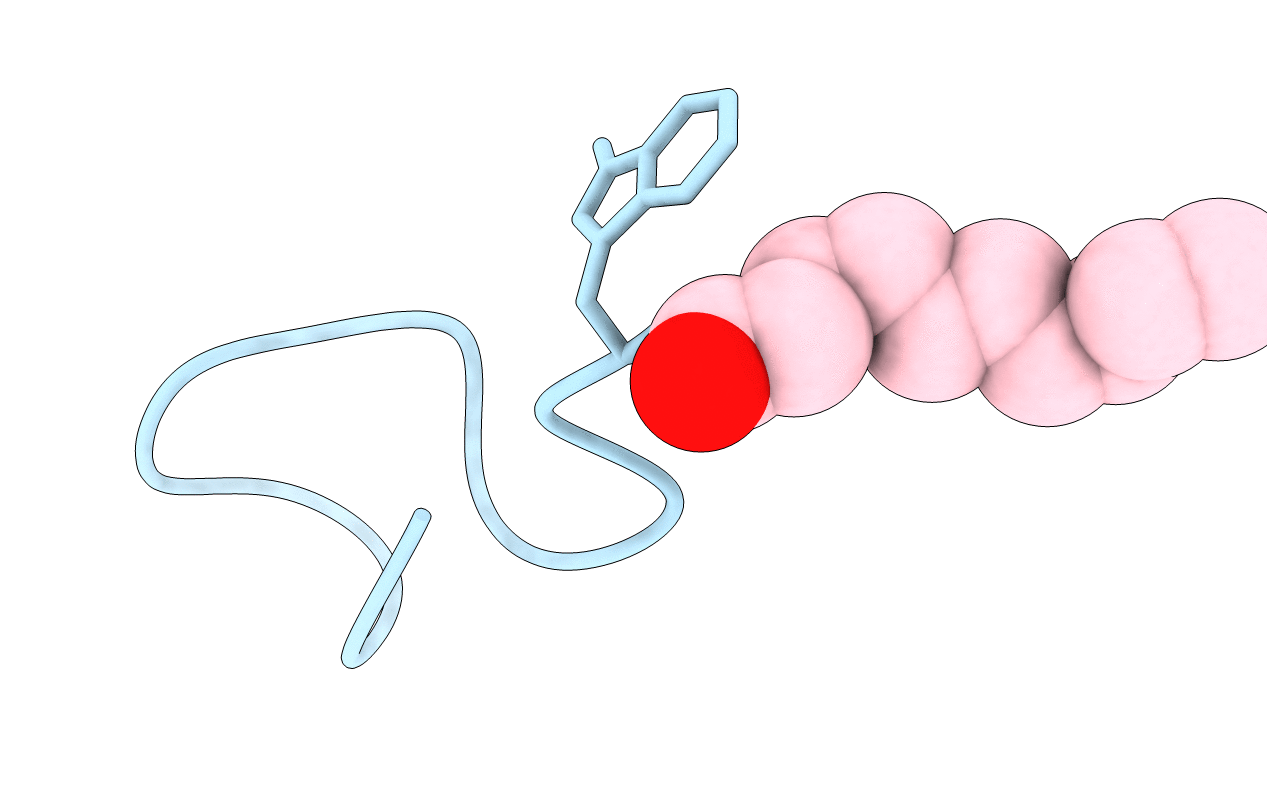 |
Organism: Streptomyces roseosporus
Method: SOLUTION NMR Release Date: 2004-11-16 Classification: ANTIBIOTIC Ligands: DKA |

