Search Count: 9
 |
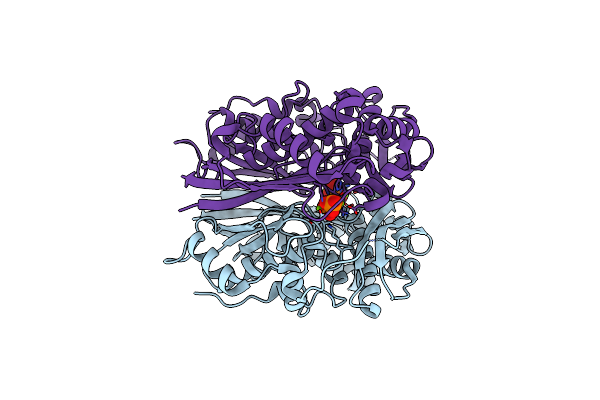
Crystal Structure Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis In Complex With Onb-Sam (2-Nitro Benzyme S-Adenosyl-Methionine)
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: 3PO, MG, EU9 |
 |
Crystal Structure Of Apo Form Of I122A/I330A Variant Of S-Adenosylmethionine Synthetase From Cryptosporidium Hominis
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-10-21 Classification: TRANSFERASE Ligands: MG, PO4 |
 |
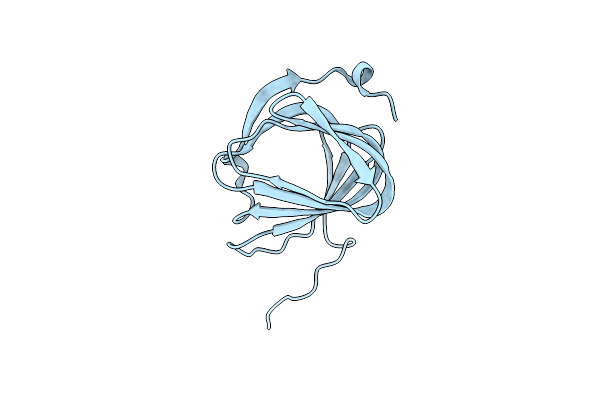
Roll Out The Beta-Barrel: Structure And Mechanism Of Pac13, A Unique Nucleoside Dehydratase
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-03-28 Classification: STRUCTURAL PROTEIN |
 |
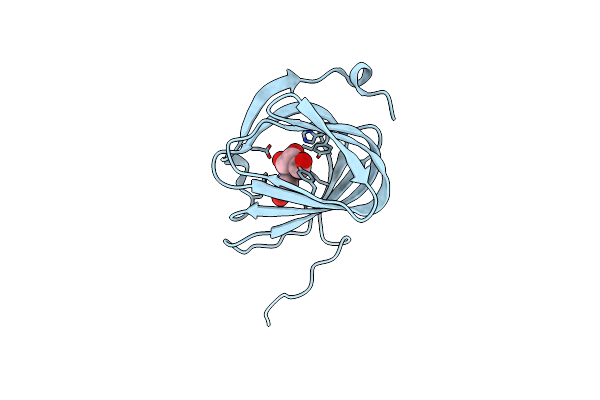
Roll Out The Beta-Barrel: Structure And Mechanism Of Pac13, A Unique Nucleoside Dehydratase
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-03-28 Classification: STRUCTURAL PROTEIN |
 |
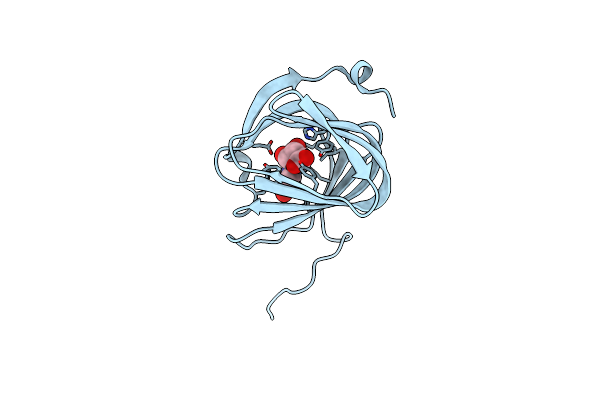
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
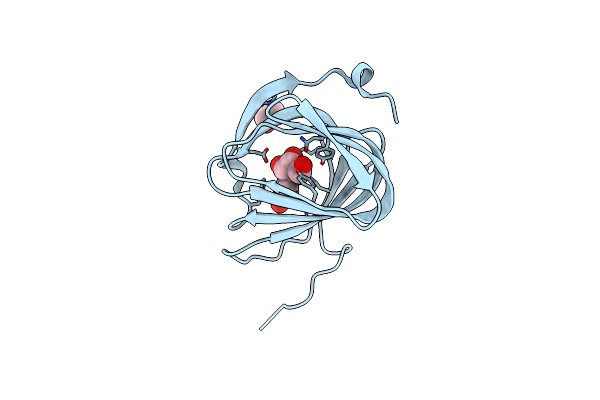
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: UUA |
 |
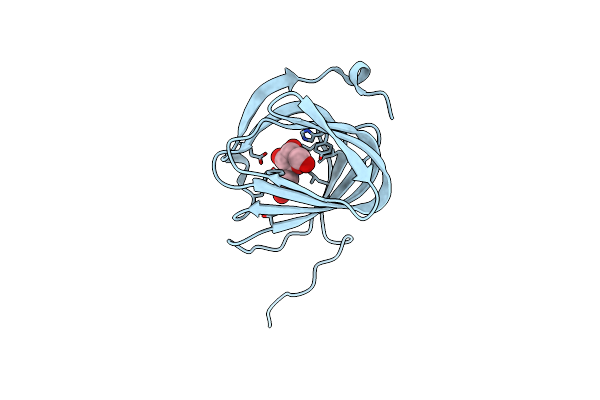
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, URI |
 |
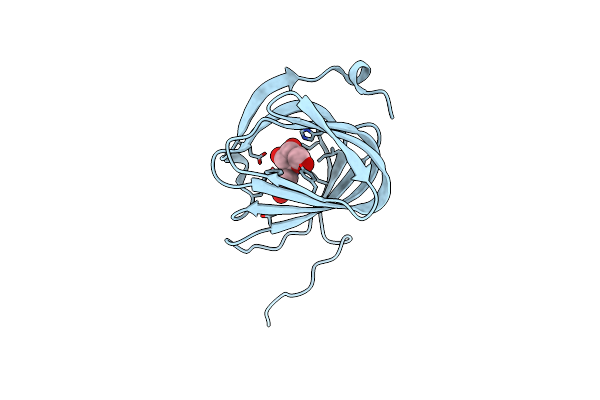
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |
 |
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-08-23 Classification: BIOSYNTHETIC PROTEIN Ligands: URI |

