Search Count: 66
 |
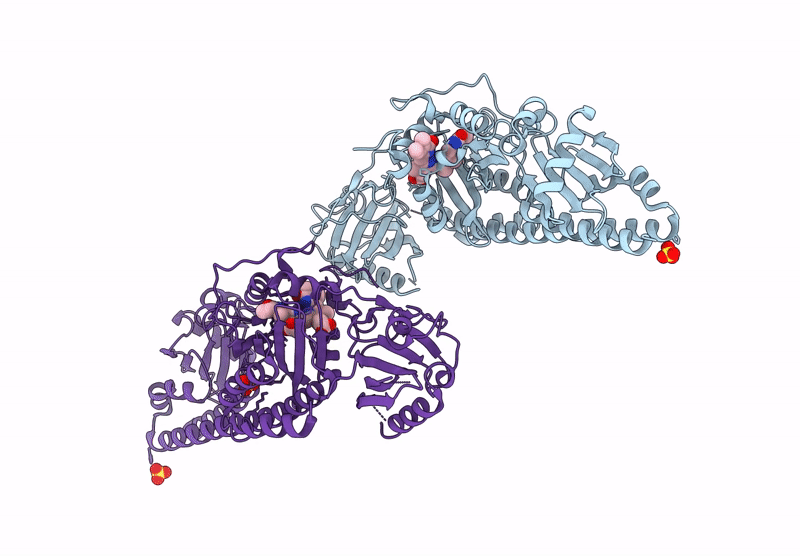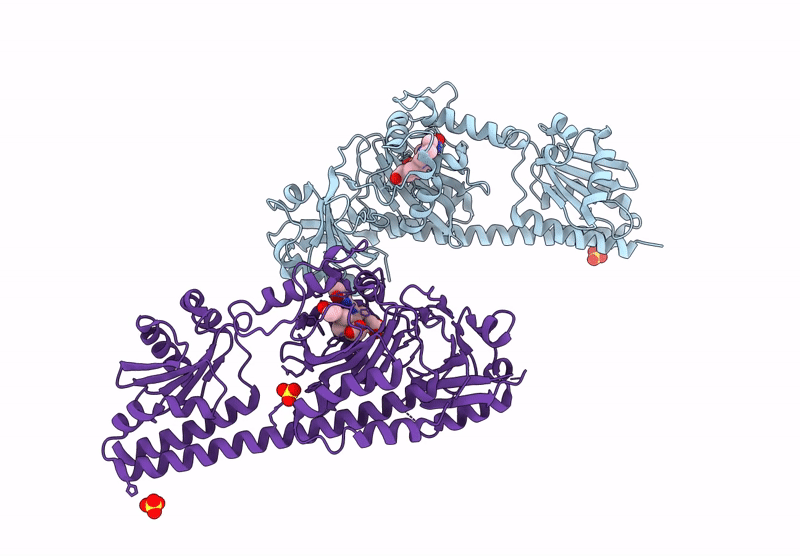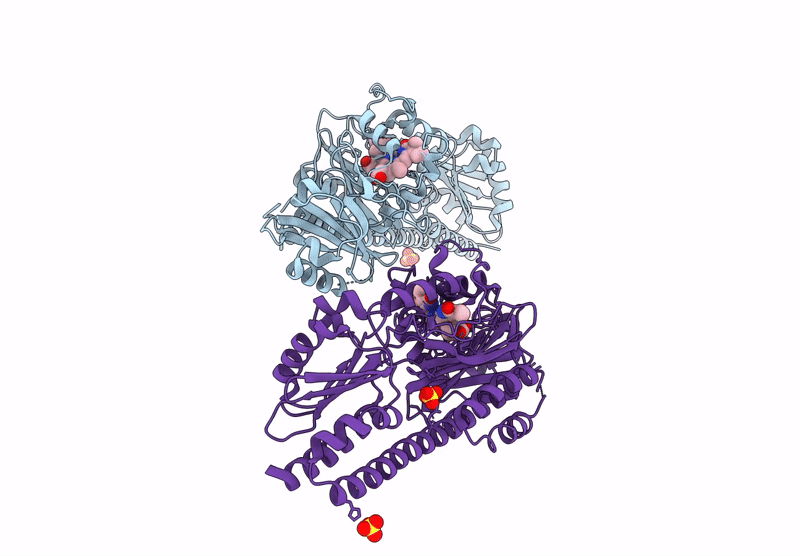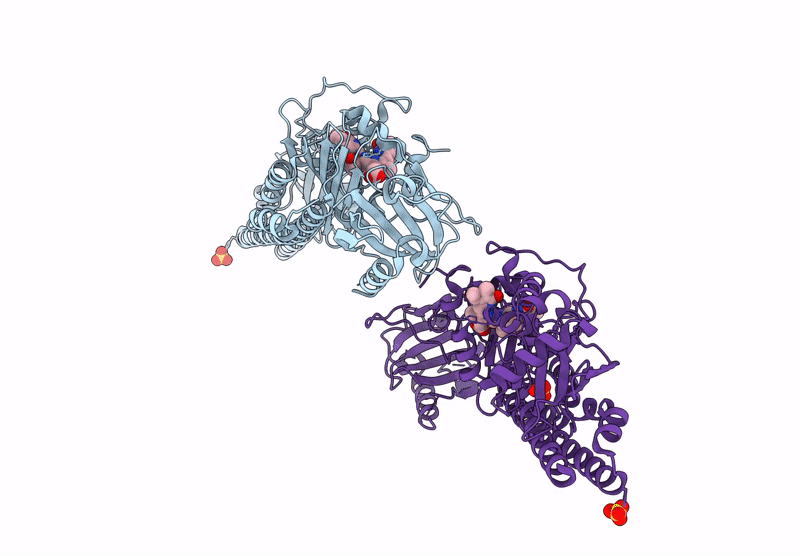
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0A).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, EDO |
 |
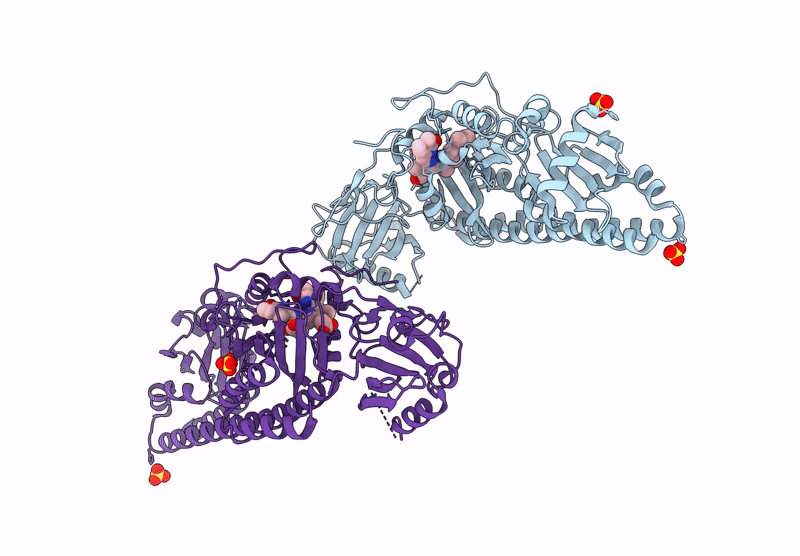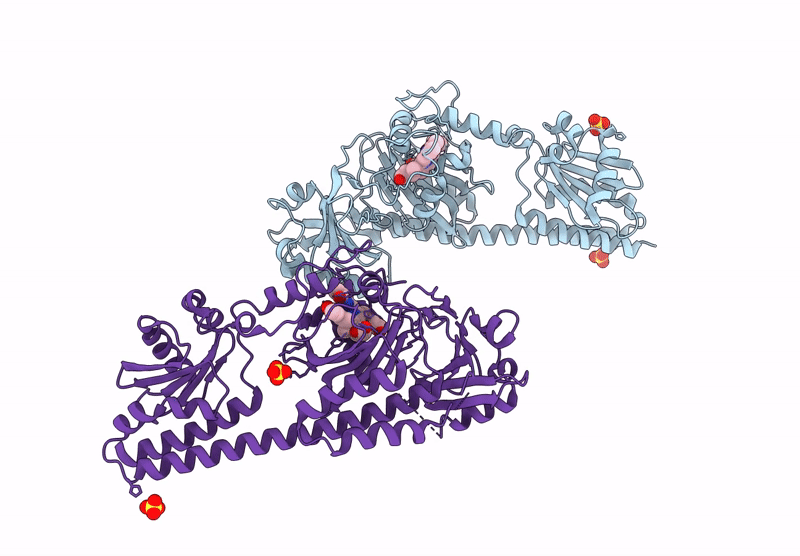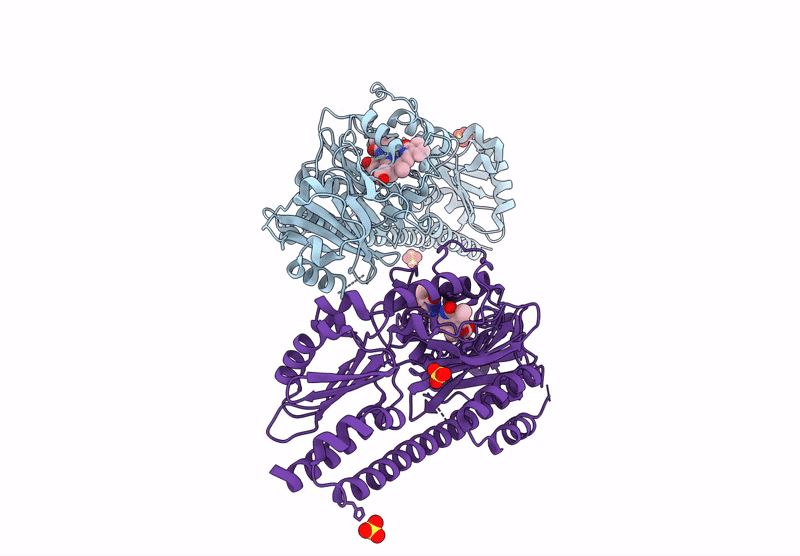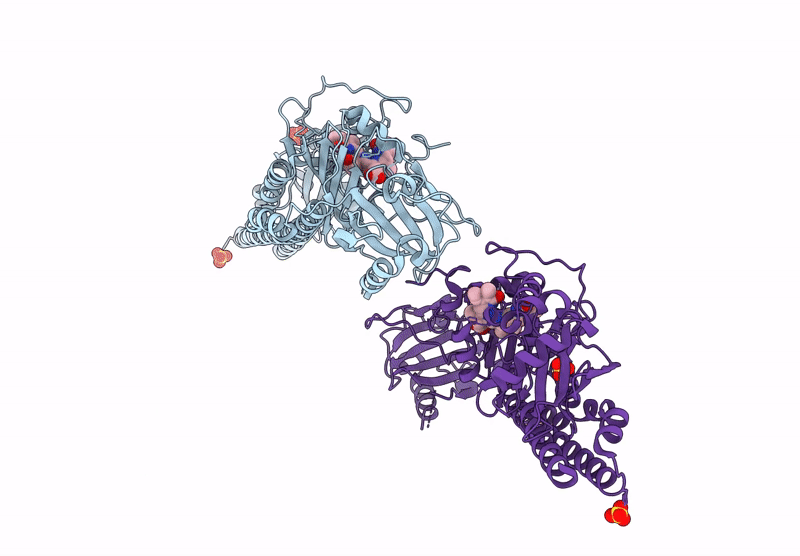
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In Its Pfr State (I0B).
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
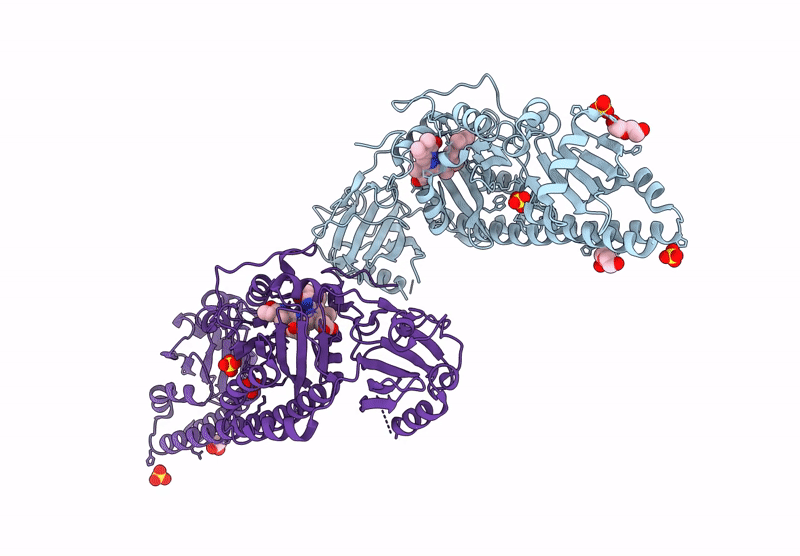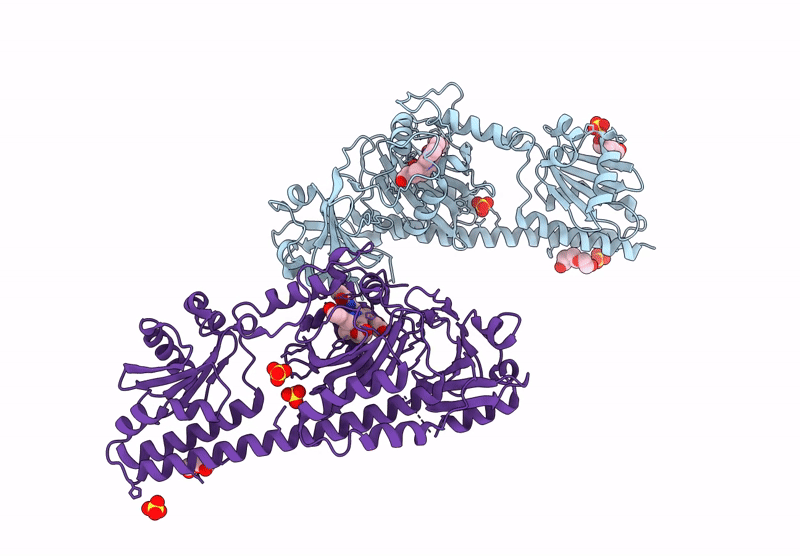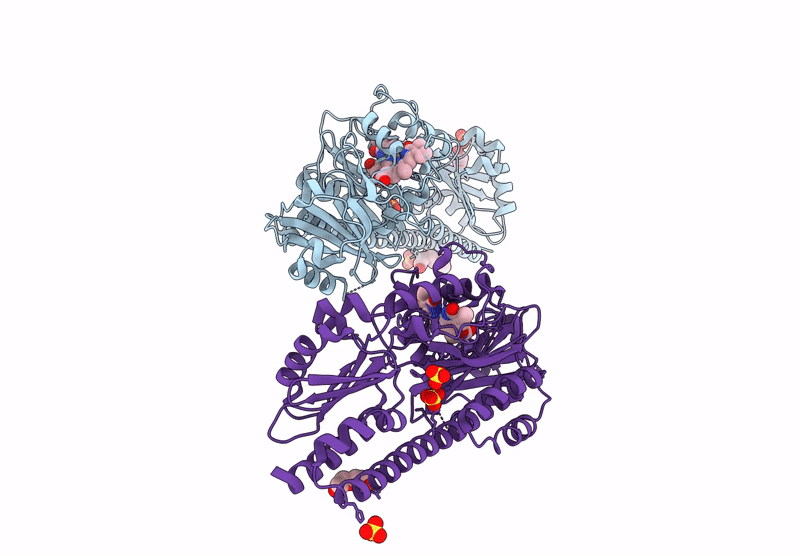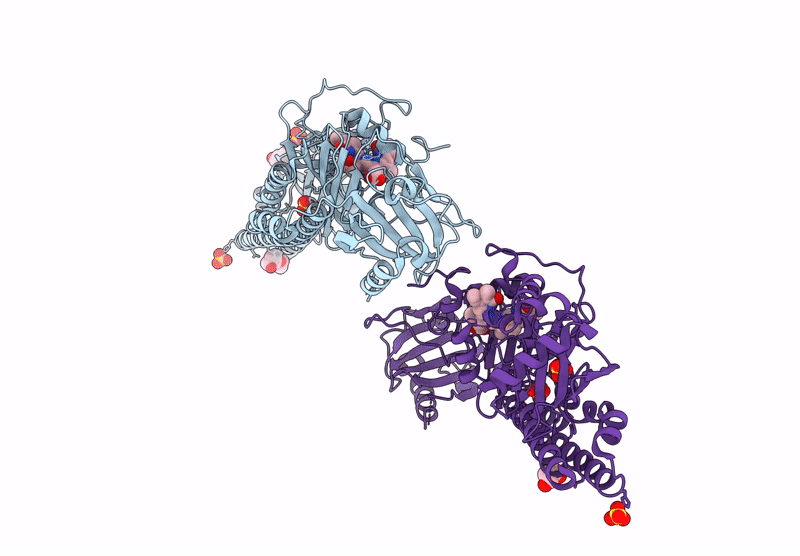
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I1 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4 |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I2 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PGE, PEG, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I3 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, GOL, PEG |
 |
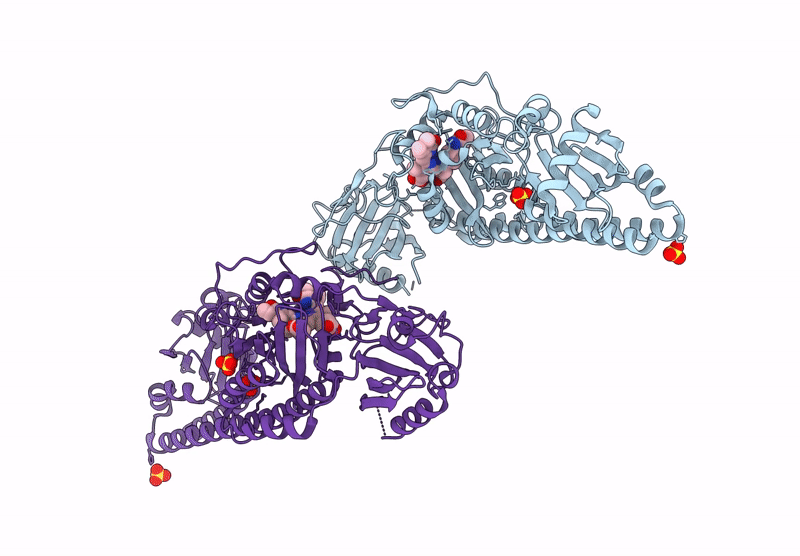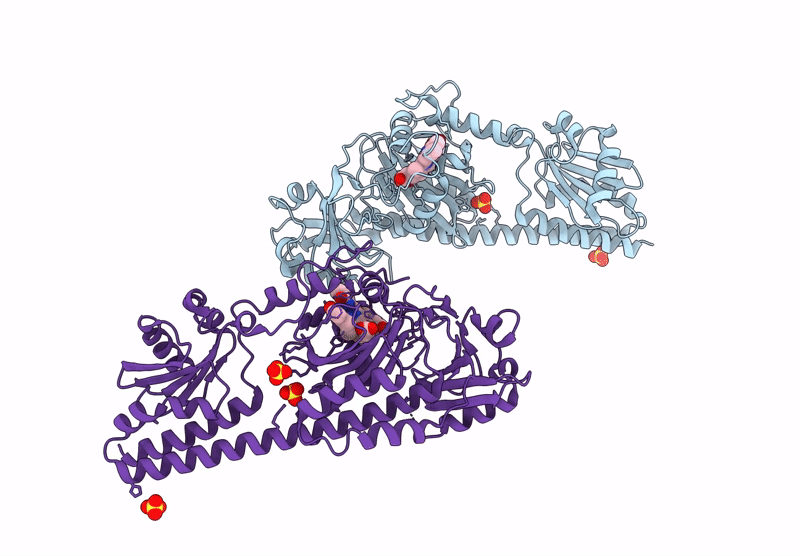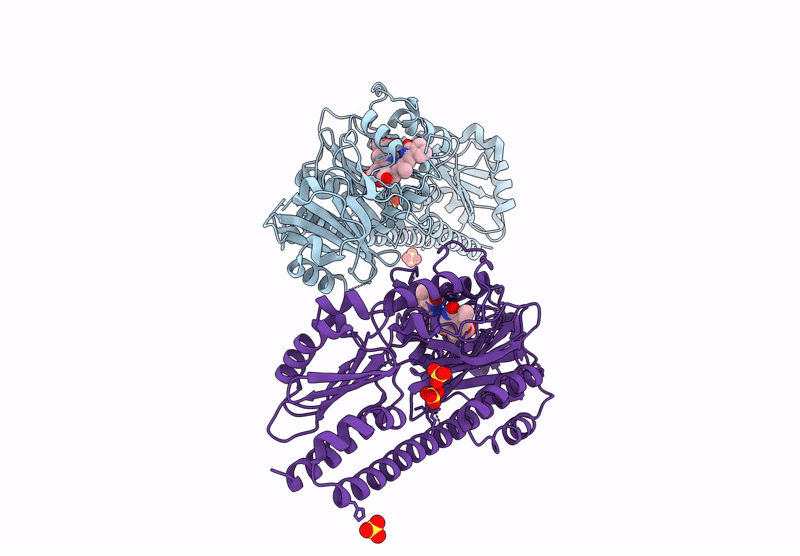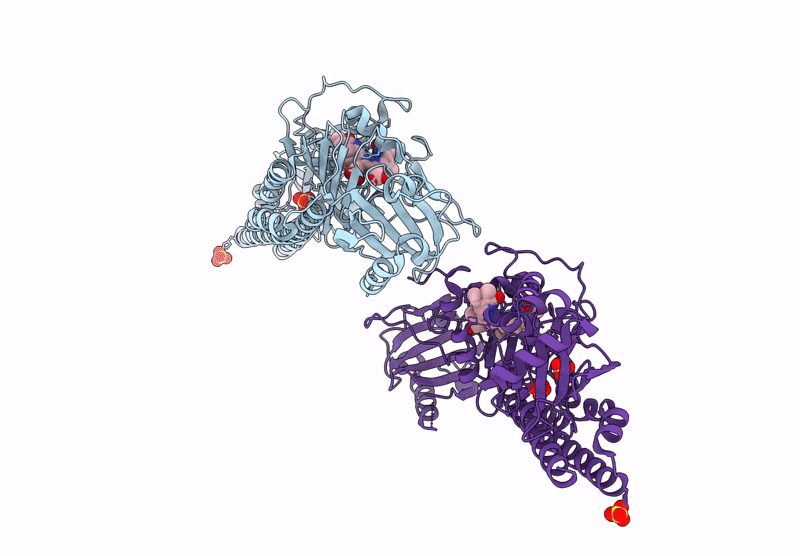
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I4 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL, PEG |
 |
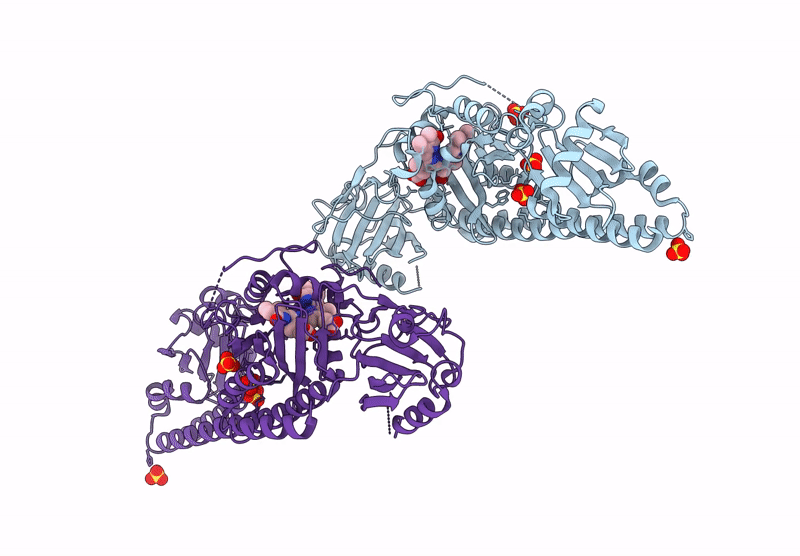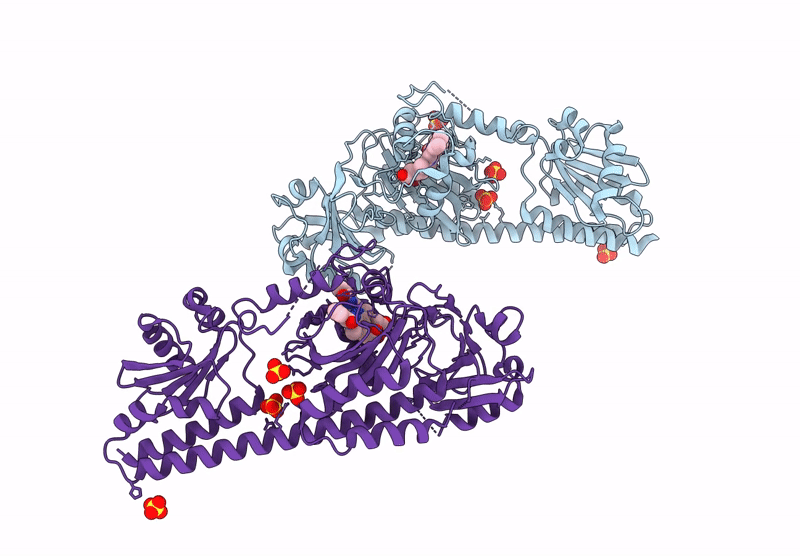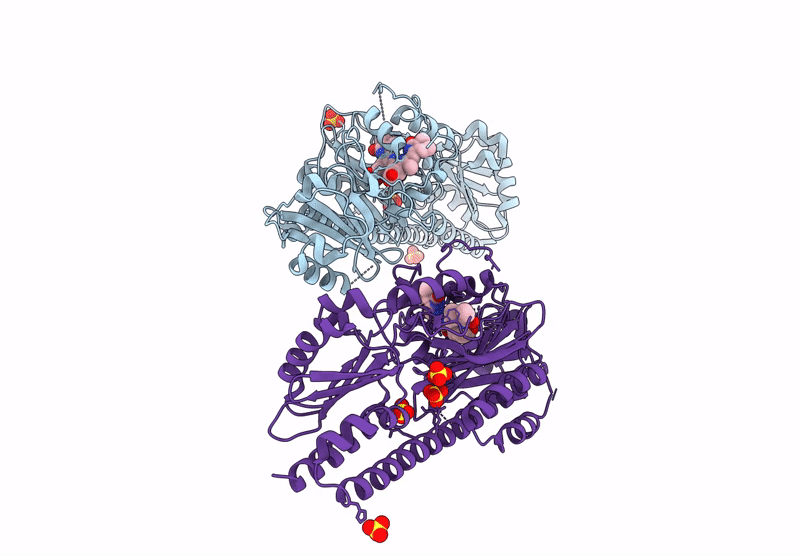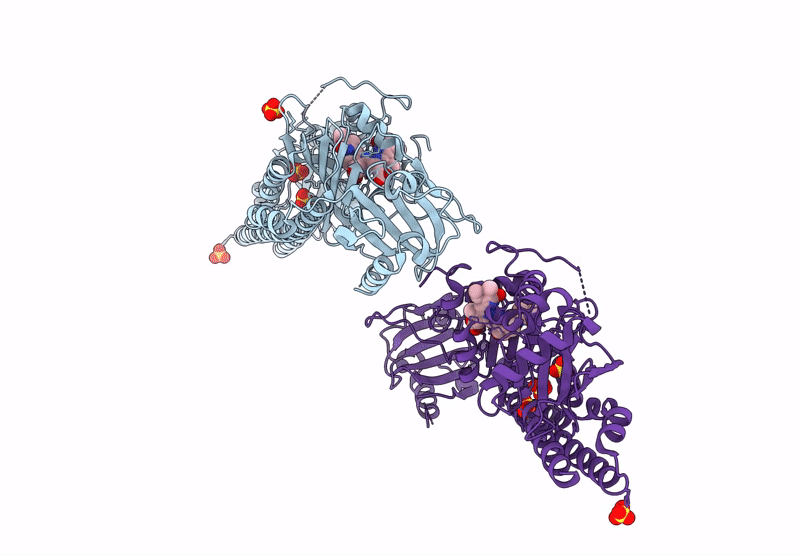
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I5 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
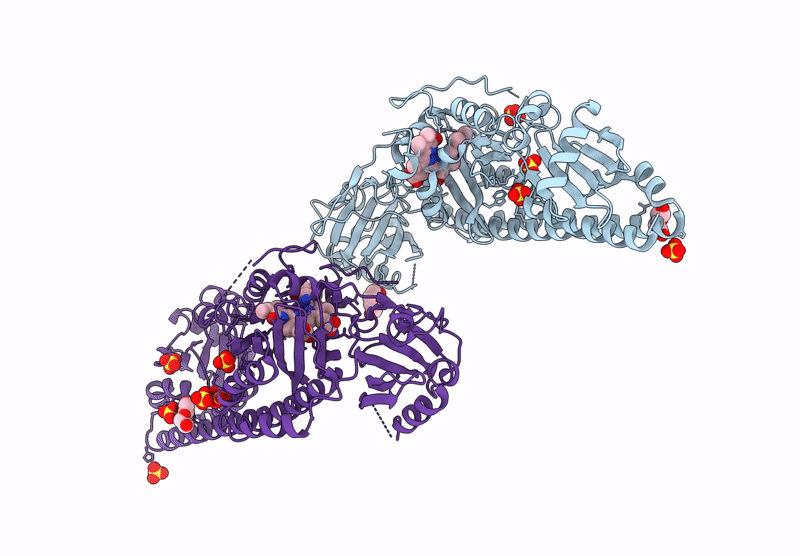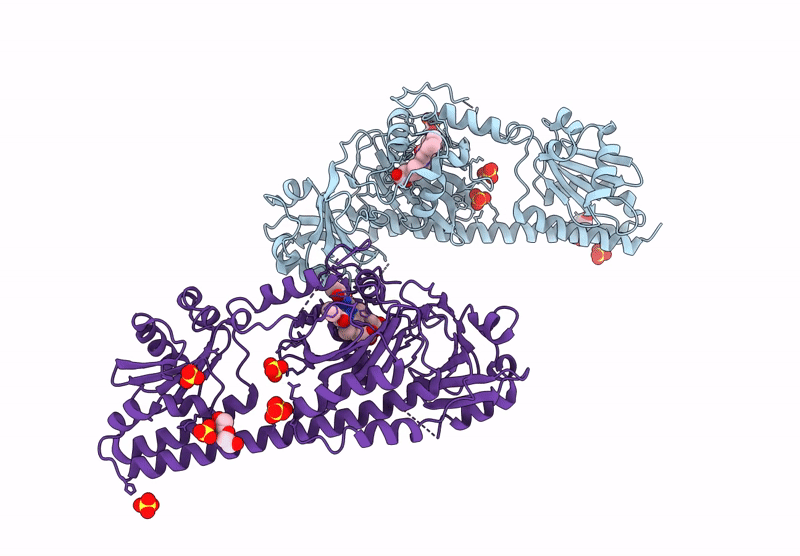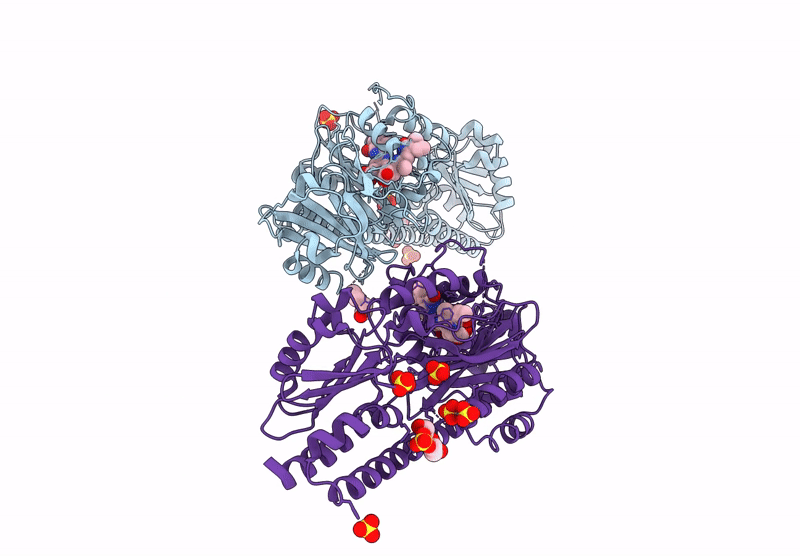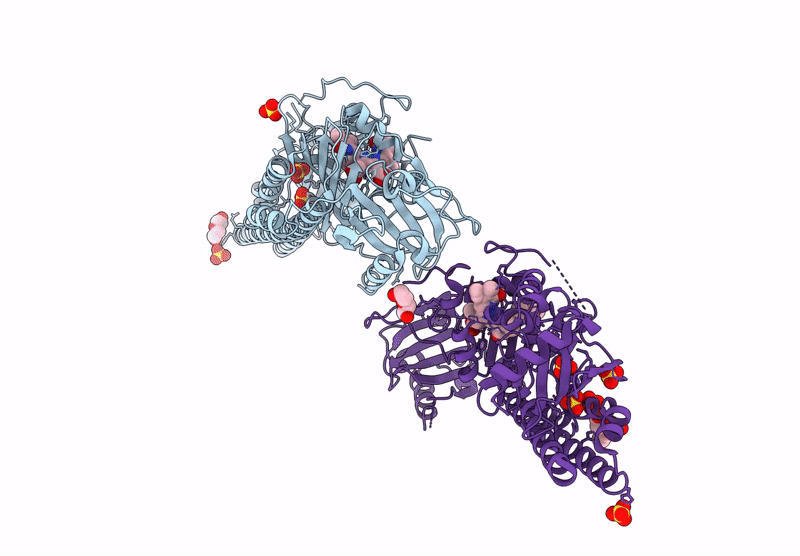
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I6 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, CL |
 |
Serial Femtosecond X-Ray Structure Of A Fluorescence Optimized Bathy Phytochrome Pairfp2 Derived From Wild-Type Agp2 In I7 Intermediate State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-14 Classification: SIGNALING PROTEIN Ligands: EL5, SO4, PEG |
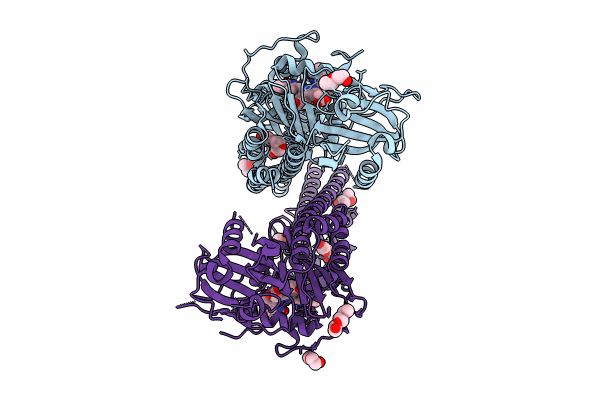 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf165 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: P6G, MPD, PEG, EL5, CL |
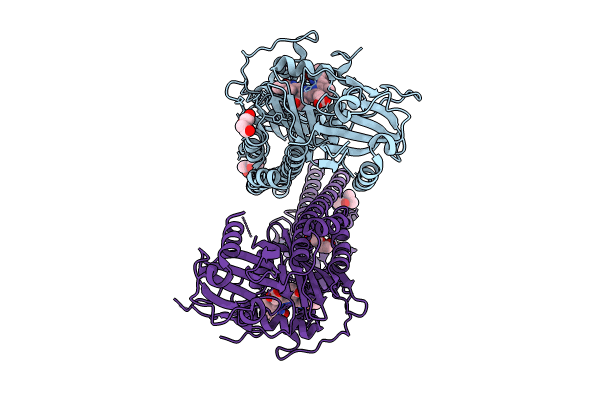 |
Crystal Structure Of The Photosensory Core Module (Pcm) Of A Cyano-Phenylalanine Mutant Ocnf192 Of The Bathy Phytochrome Agp2 From Agrobacterium Fabrum In The Pfr State.
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: PEG, MPD, EL5, CL |
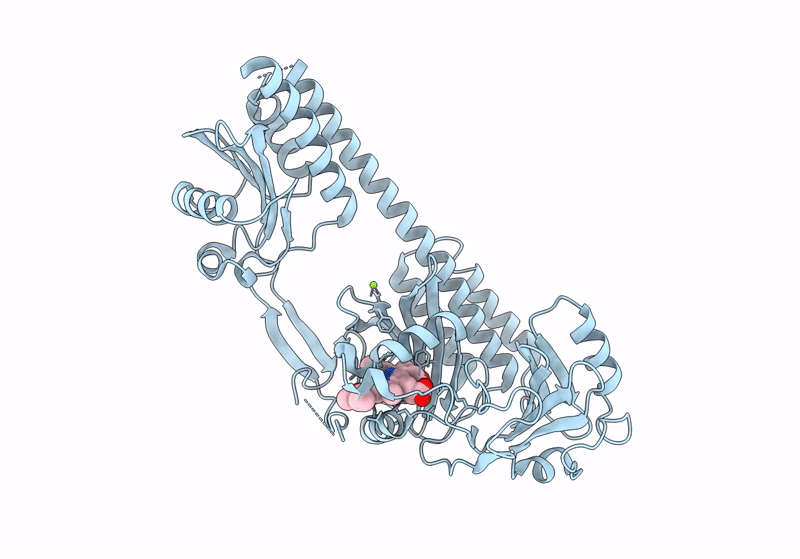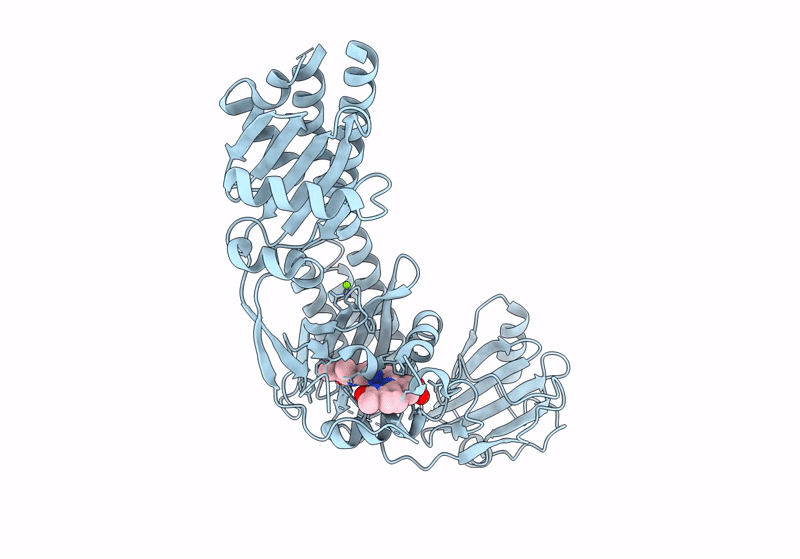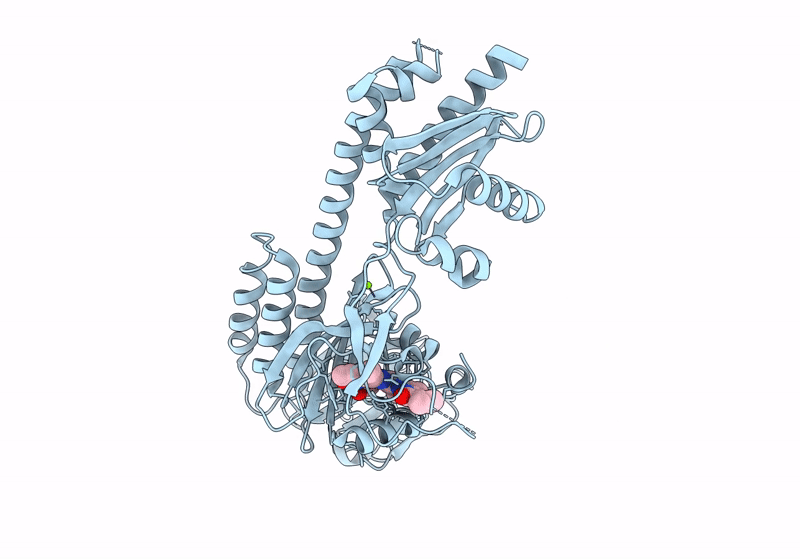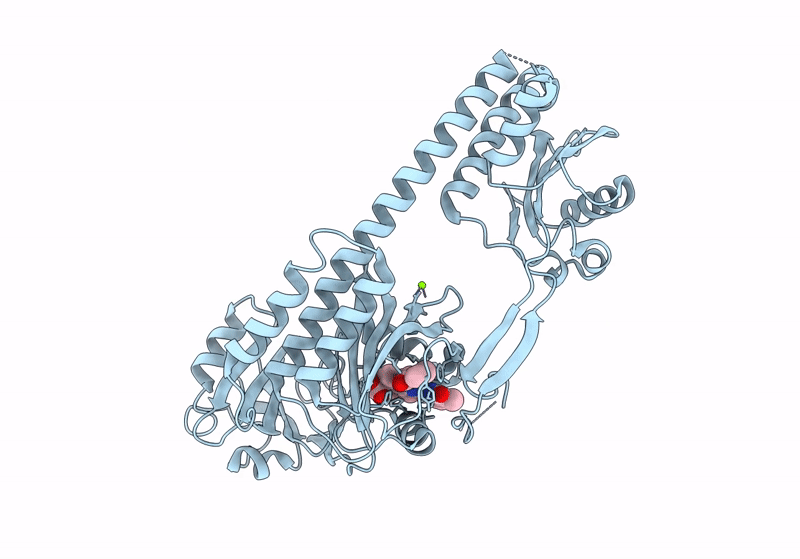 |
The Surface-Engineered Photosensory Module (Pas-Gaf-Phy) Of The Bacterial Phytochrome Agp1 (Atbphp1) In The Pr Form With Parallel Dimer Formation
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-03-06 Classification: SIGNALING PROTEIN Ligands: V8U, MG |
 |
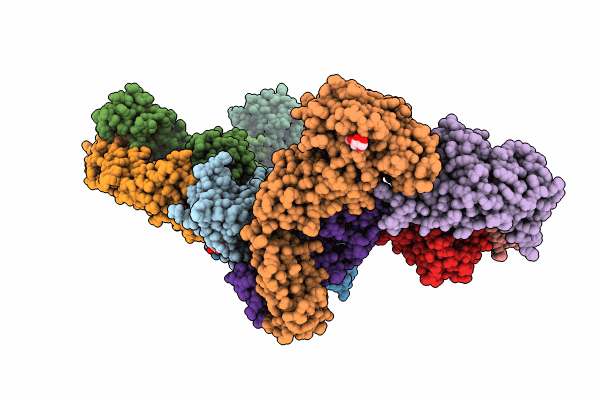
Crystal Structure Of The Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Wrair-5021
Organism: Severe acute respiratory syndrome coronavirus 2, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2024-01-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: GOL, NAG, MLI |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Wrair-5001
Organism: Severe acute respiratory syndrome coronavirus 2, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2024-01-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, GOL |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Sars-Cov-2 Reactive Human Antibody Cr3022
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:4.22 Å Release Date: 2023-12-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
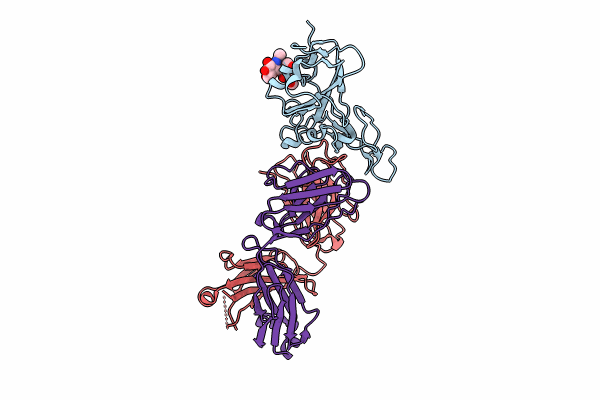
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-12-13 Classification: VIRAL PROTEIN Ligands: NAG, FUC, GOL |
 |
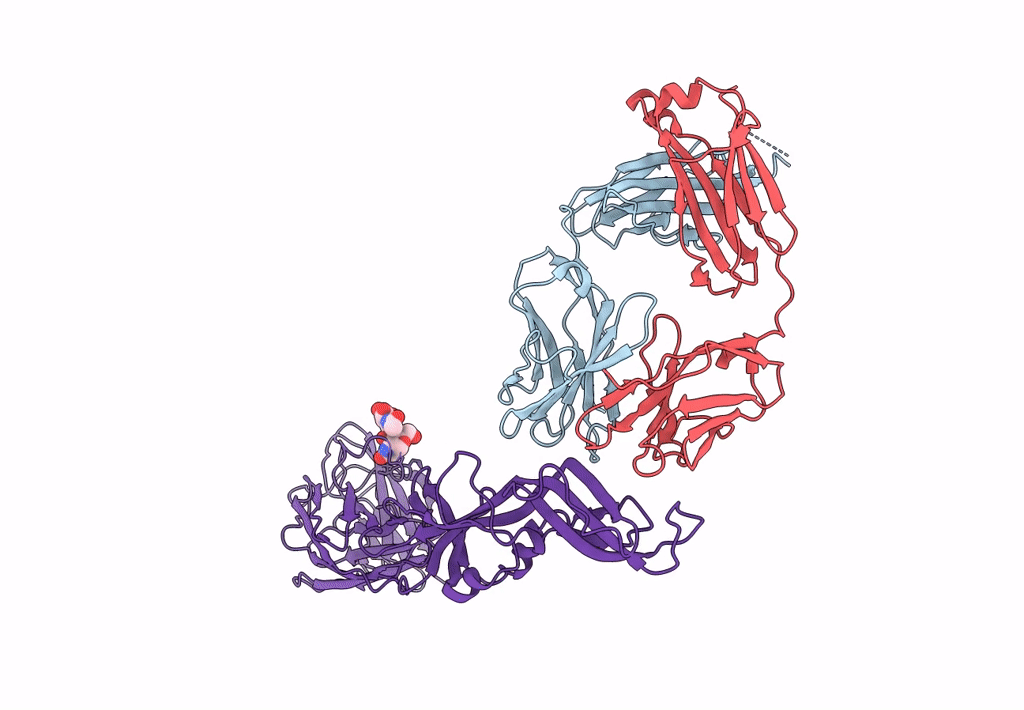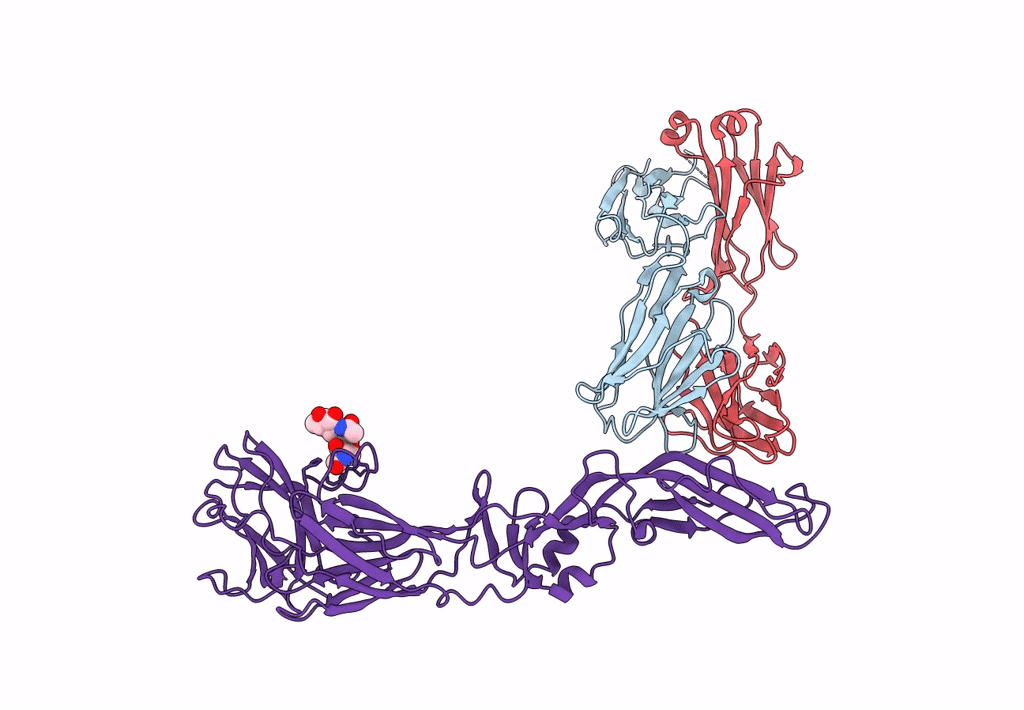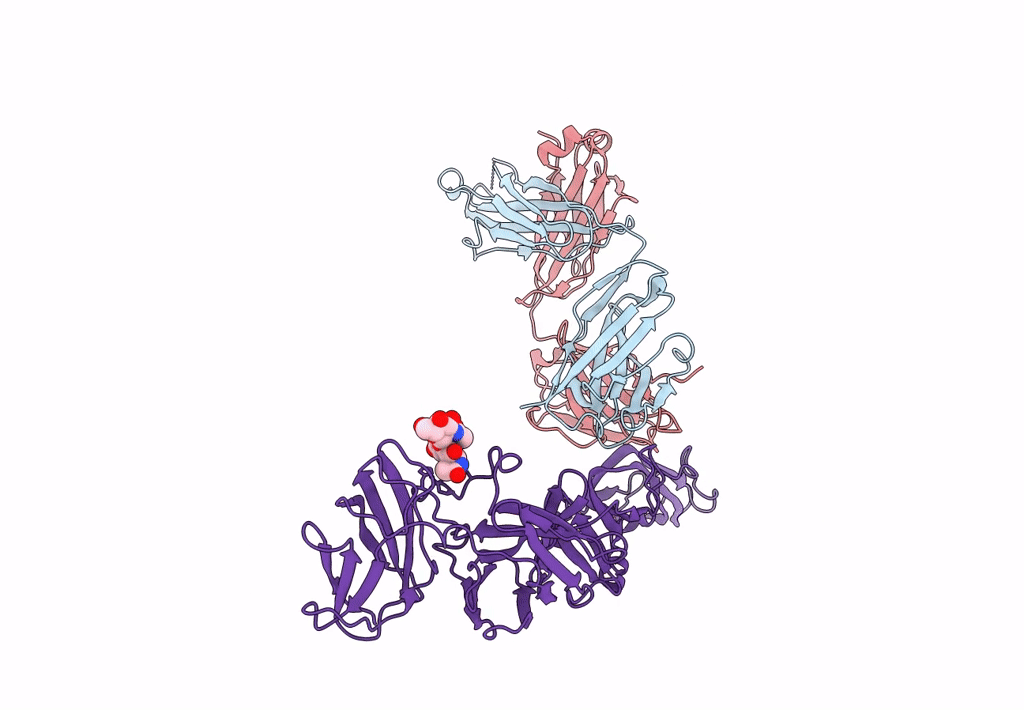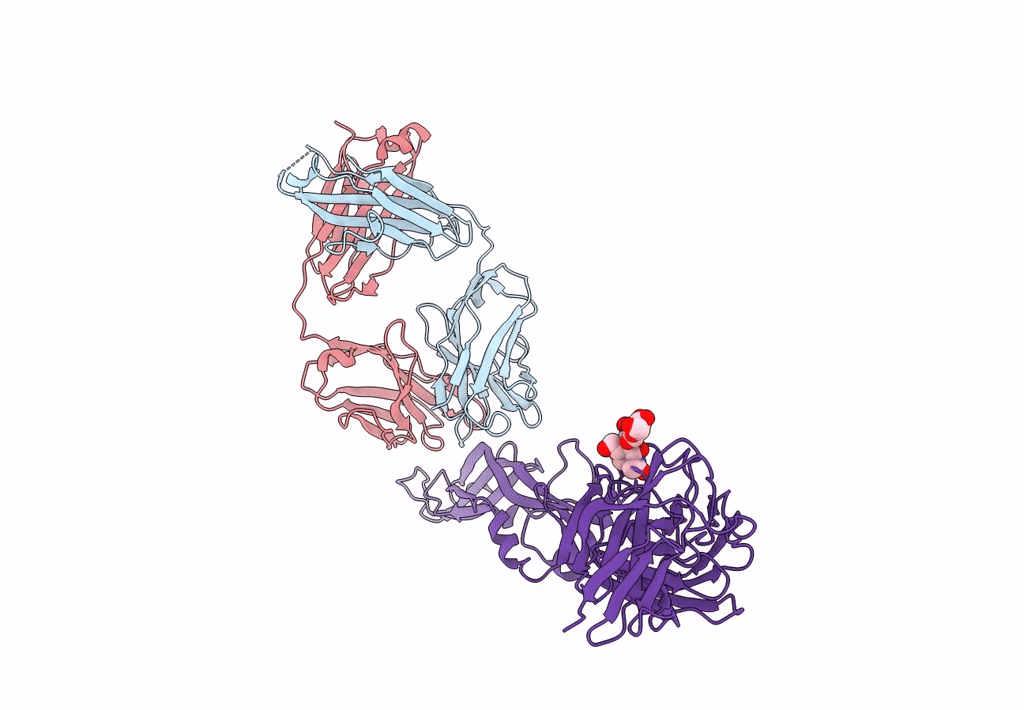
Crystal Structure Of Antibody Wrair-2123 In Complex With Sars-Cov-2 Receptor Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-12-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of A Nhp Anti-Zikv Neutralizing Antibody Rhmz119-D In Complex With Zikv E Glycoprotein
Organism: Zika virus zikv/h. sapiens/frenchpolynesia/10087pf/2013, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:3.58 Å Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of A Nhp Anti-Zikv Neutralizing Antibody Rhmz100-C In Complex With Zikv E Glycoprotein
Organism: Zika virus zikv/h. sapiens/frenchpolynesia/10087pf/2013, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of A Nhp Anti-Zikv Neutralizing Antibody Rhmz107-B In Complex With Zikv E Glycoprotein
Organism: Zika virus zikv/h. sapiens/frenchpolynesia/10087pf/2013, Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2023-08-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

