Search Count: 14
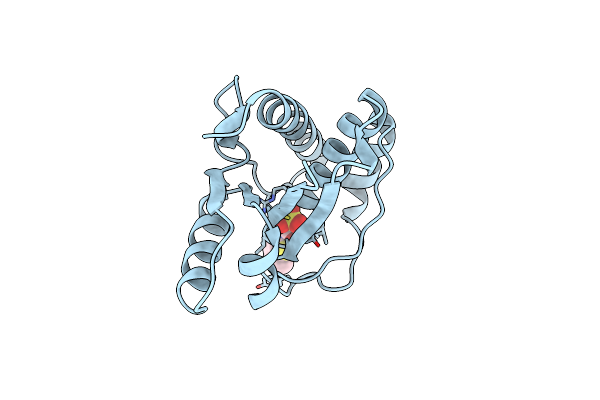 |
The Co-Crystal Structure Of Low Molecular Weight Protein Tyrosine Phosphatase (Lmw-Ptp) With A Small Molecule Inhibitor Spaa-2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-10-19 Classification: HYDROLASE Ligands: OIF |
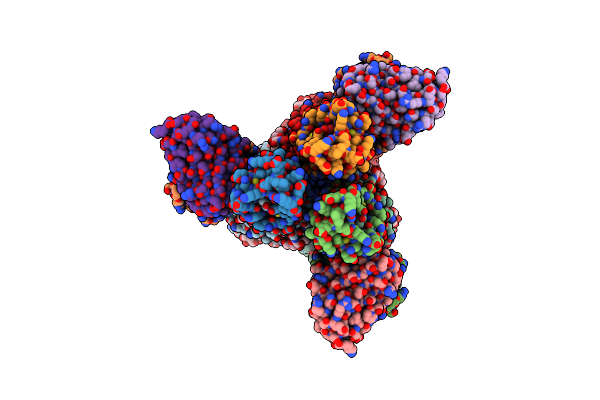 |
Organism: Influenza a virus (strain a/new zealand:south canterbury/35/2000 h1n1), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
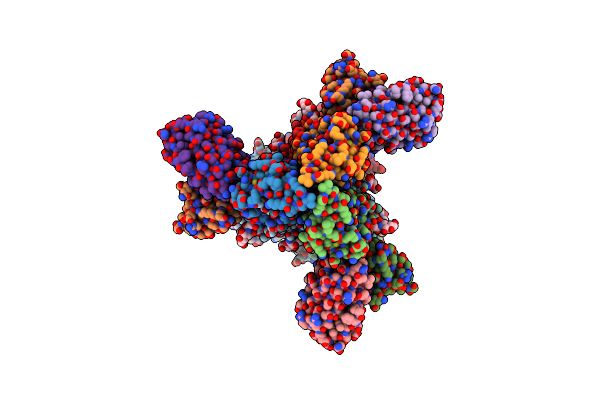 |
Organism: Influenza a virus (strain a/new zealand:south canterbury/35/2000 h1n1), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
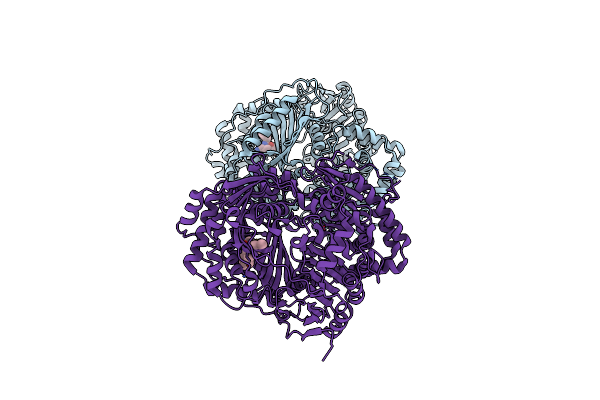 |
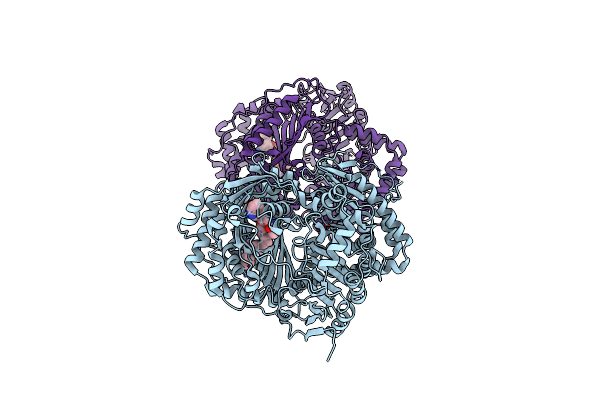
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2015-06-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2QW, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-06-17 Classification: Hydrolase/Hydrolase Inhibitor Ligands: 2Q6, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2015-06-17 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2QX, ZN |
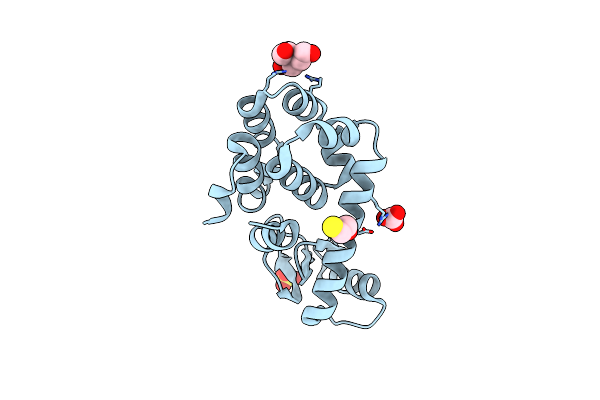 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Apo Structure
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: BME, CO3, TAM, SO4 |
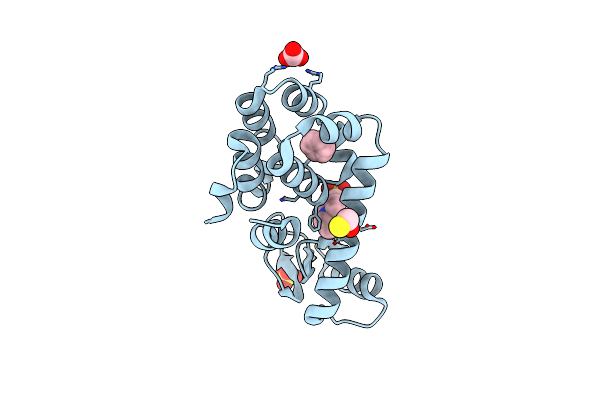 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Benzene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: BNZ, BME, CO3, EPE, SO4 |
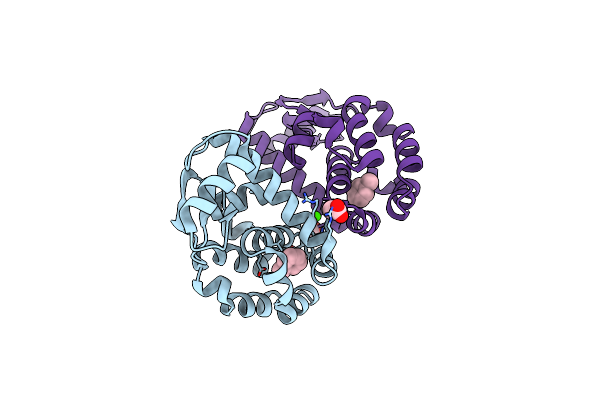 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Toluene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: MBN, CA, CL, ACT |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Ethylbenzene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: PYJ, ACT, CA, CL |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--P-Xylene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: PXY, CL, ACT, CA |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Aniline Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: ANL, CA, CL |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Phenol Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: IPH, CL, CA |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Pyridine Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: 0PY, CO3, MG, CL |

