Search Count: 16
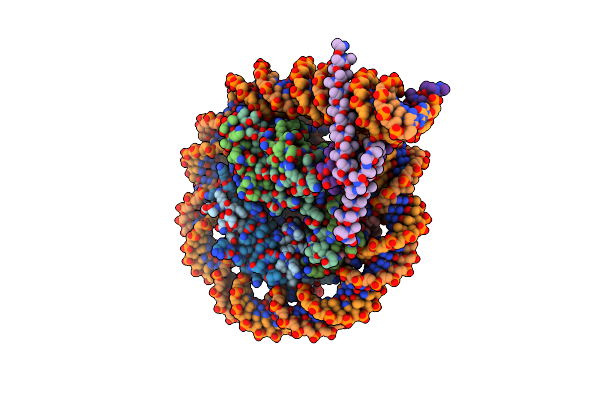 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl-6.2 (Dna Conformation 1)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
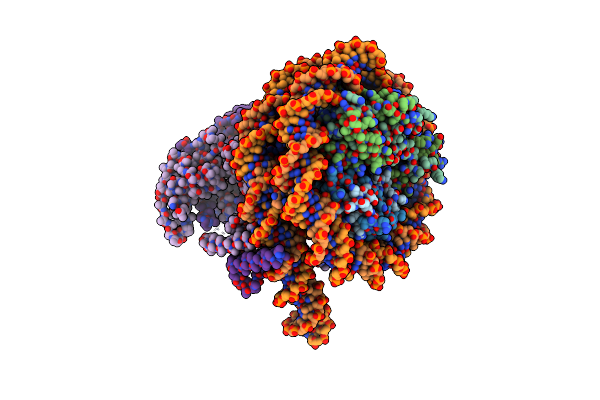 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl+5.8 (Composite Map)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
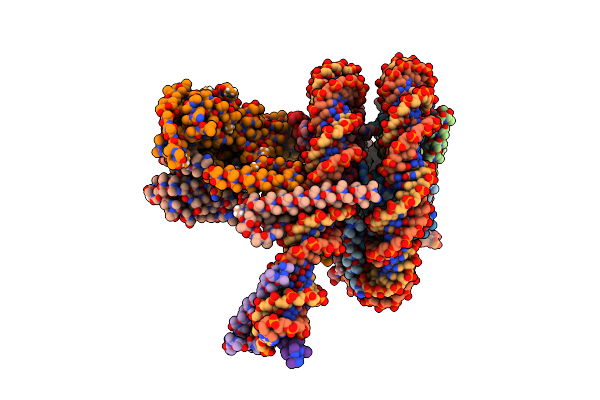 |
Cryo-Em Structure Of Clock-Bmal1 Bound To The Native Por Enhancer Nucleosome (Map 2, Additional 3D Classification And Flexible Refinement)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
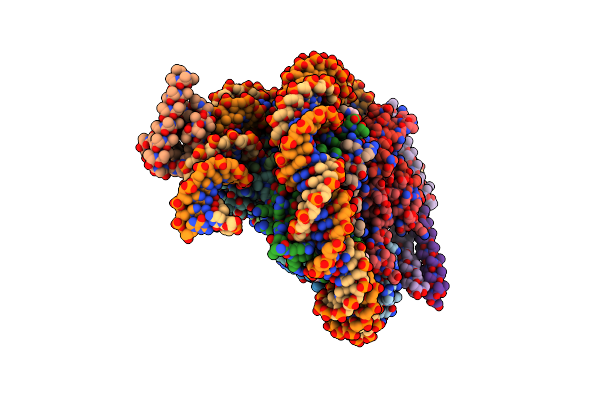 |
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-06 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-06 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-06 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2020-03-04 Classification: CIRCADIAN CLOCK PROTEIN |
 |
Crystal Structure Of Foldswitch-Stabilized Kaib In Complex With The N-Terminal Ci Domain Of Kaic From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus, Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-29 Classification: Transcription Regulator Ligands: ADP |
 |
Crystal Structure Of Kaic S431E In Complex With Foldswitch-Stabilized Kaib From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:3.87 Å Release Date: 2017-03-29 Classification: Transcription Regulator Ligands: ADP |
 |
Crystal Structure Of Foldswitch-Stabilized Kaib In Complex With The N-Terminal Ci Domain Of Kaic And A Dimer Of Kaia C-Terminal Domains From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus, Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2017-03-29 Classification: Transcription Regulator Ligands: PO4 |
 |
Nmr Structure Of Foldswitch-Stablized Kaib From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus
Method: SOLUTION NMR Release Date: 2017-03-29 Classification: Sigaling protein |
 |
Nmr Structure Of Pseudo Receiver Domain Of Cika From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: SOLUTION NMR Release Date: 2017-03-29 Classification: Sigaling protein |
 |
Nmr Structure Of Foldswitch-Stablized Kaib In Complex With Pseudo Receiver Domain Of Cika From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus (strain bp-1), Thermosynechococcus elongatus
Method: SOLUTION NMR Release Date: 2017-03-29 Classification: Sigaling protein |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-02-08 Classification: TRANSCRIPTION Ligands: CL |

