Search Count: 526
 |
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
The Cryo-Em Structure Of Porcine Serum Mgam Bound With Acarviosyl-Maltotriose.
Organism: Sus scrofa domesticus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: HYDROLASE Ligands: NAG |
 |
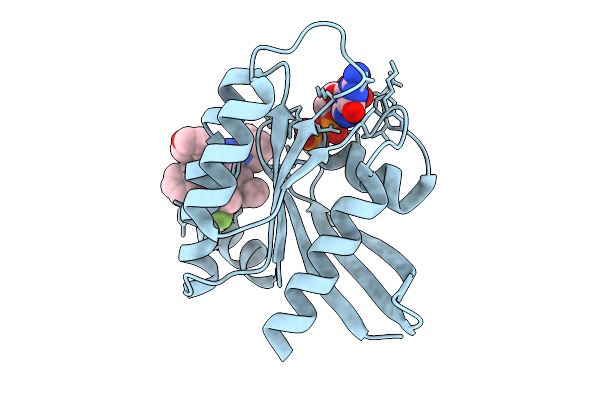
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: EDO, CIT |
 |
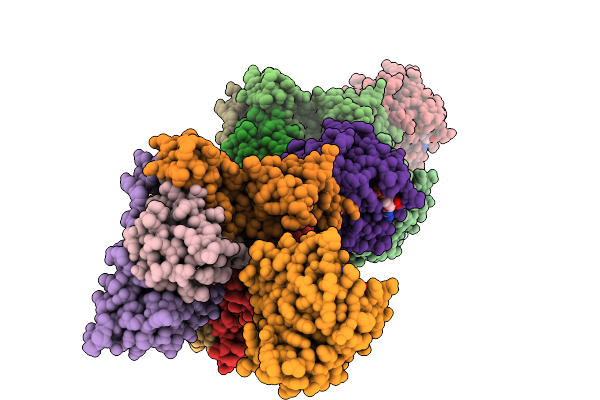
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: GDP, MG, A1L65 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: A1L66, GDP, MG |
 |
Crystal Structure Of Hla-C*12:02 In Complex With Kaynvtqaf (Kf9), A 9-Mer Epitope From Sars-Cov-2 Nucleocapsid (N266-274)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Gv37-Tcr In Complex With Hla-C*12:02 With Kaynvtqaf (Kf9), A 9-Mer Epitope From Sars-Cov-2 Nucleocapsid (N266-274)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: IMMUNE SYSTEM Ligands: EDO, SO4 |
 |
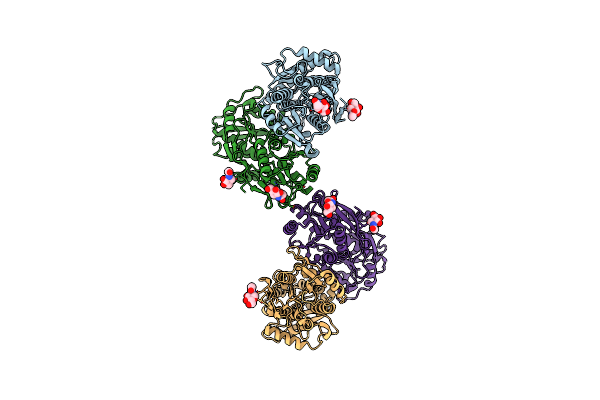
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: NUCLEAR PROTEIN Ligands: A1MAM, EDO, ACT |
 |
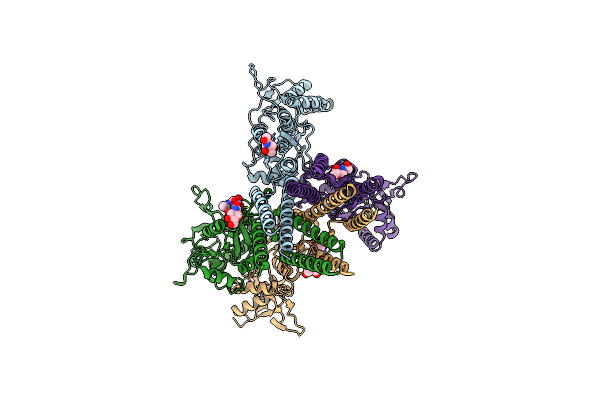
Structure Of The Amino-Terminal Domain Of Kainate Receptor Gluk2 In The Apo State
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Structure Of Ligand Binding And Transmembrane Domains Of Kainate Receptor Gluk2 In Apo State
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Kalium Channelrhodopsin 1 C110A Mutant From Hyphochytrium Catenoides, Dark State
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PEE |
 |
Kalium Channelrhodopsin 1 C110A Mutant From Hyphochytrium Catenoides, Laser-Flash-Illuminated
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PEE |
 |
Kalium Channelrhodopsin 1 C110A Mutant From Hyphochytrium Catenoides, Continuous Illumination State
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: MEMBRANE PROTEIN Ligands: RET, CLR, PEE |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION |
 |
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-04 Classification: HYDROLASE Ligands: IB0 |
 |
Crystal Structure Of Thermus Thermophilus Peptidyl-Trna Hydrolase In Complex With Adenosine 5'-Monophosphate
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2024-11-20 Classification: TRANSLATION Ligands: AMP, TLA, GOL |

