Search Count: 517
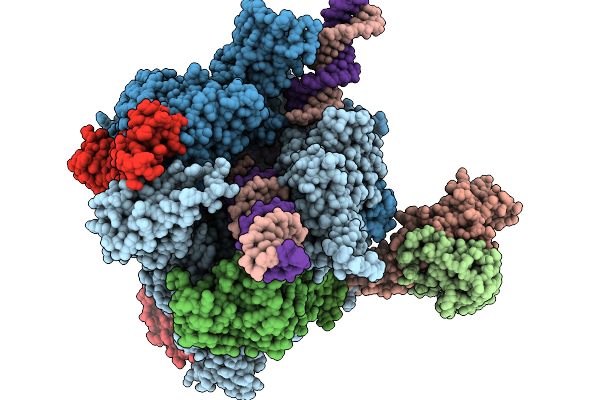 |
Structure Of A Native Drosophila Melanogaster Pol Ii Elongation Complex With A Well-Defined Rpb4/Rpb7 Stalk
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Resolution:3.44 Å Release Date: 2026-01-07 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
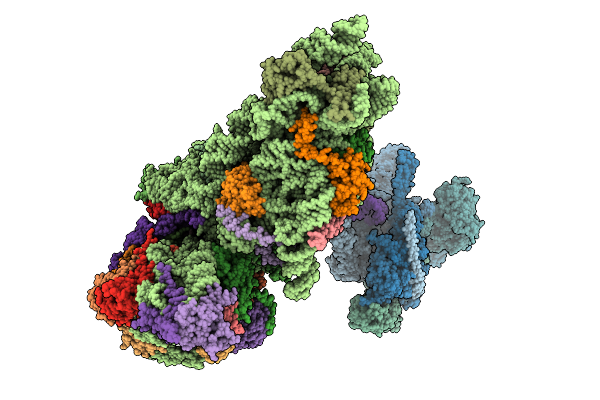
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
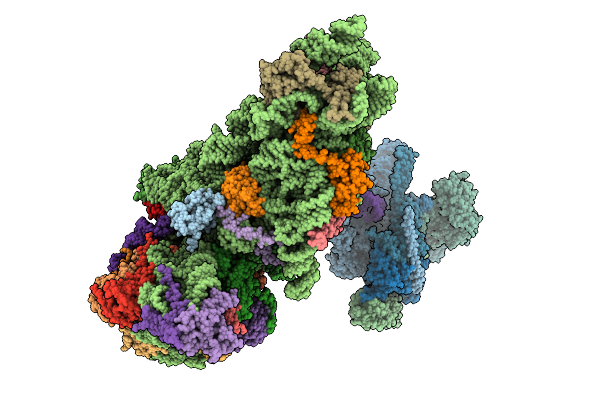
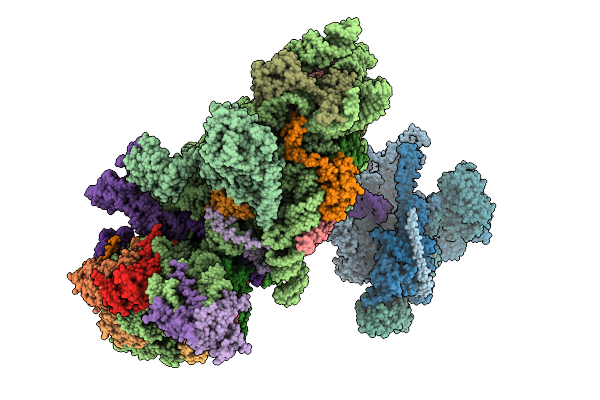
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
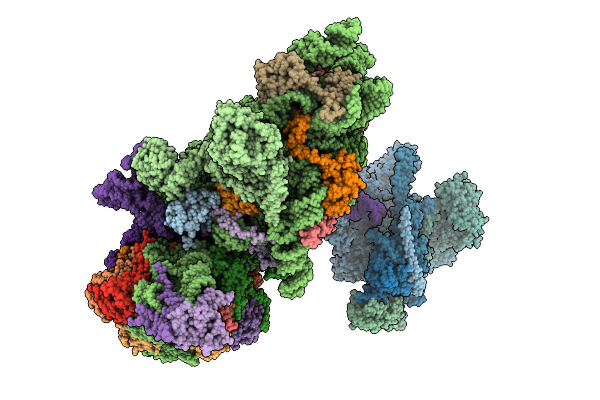
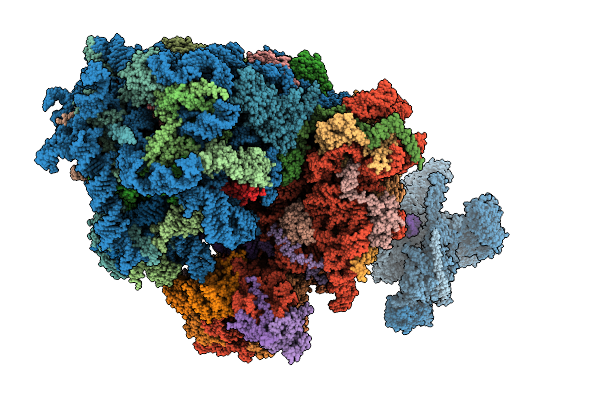
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
 |
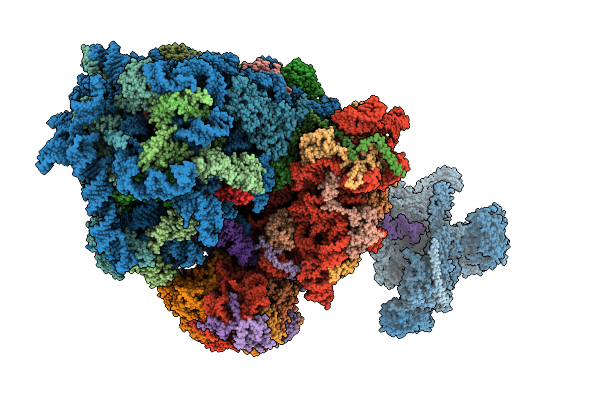
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
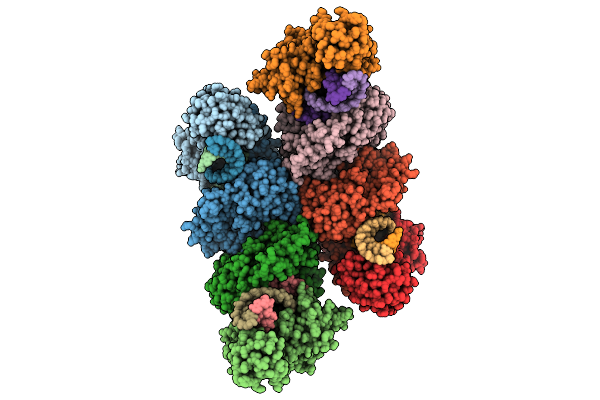
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
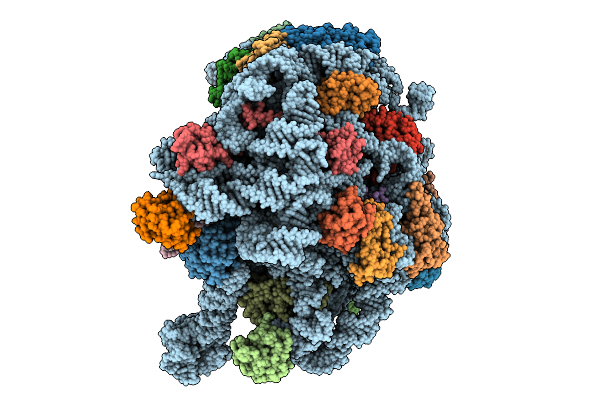
Cryo-Em Structure Of Filament Form Acidithiobacillus Caldus (Aca) Short Prokaryotic Argonautes, Hnh-Associated (Sparha) With Grna And Tdna
Organism: Acidithiobacillus caldus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: IMMUNE SYSTEM/DNA/RNA Ligands: MN |
 |
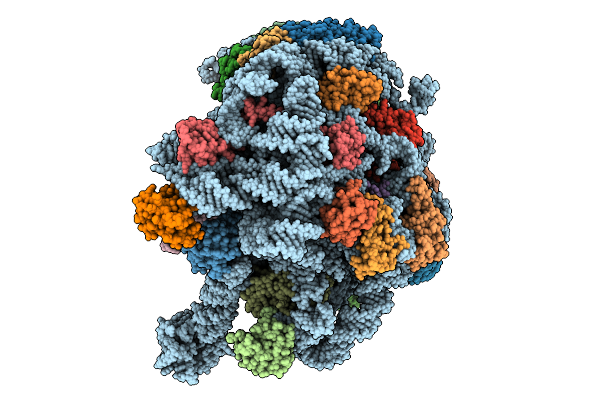
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, K, ZN |
 |
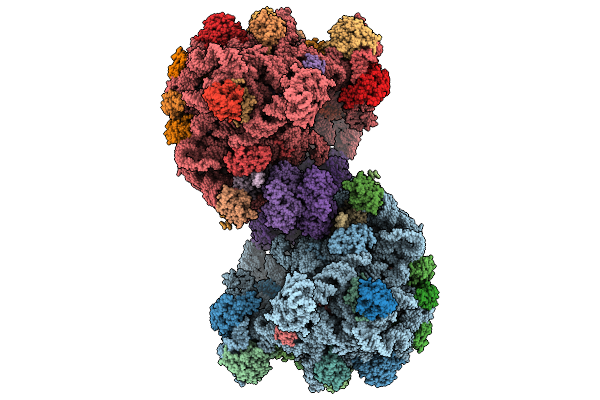
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME |
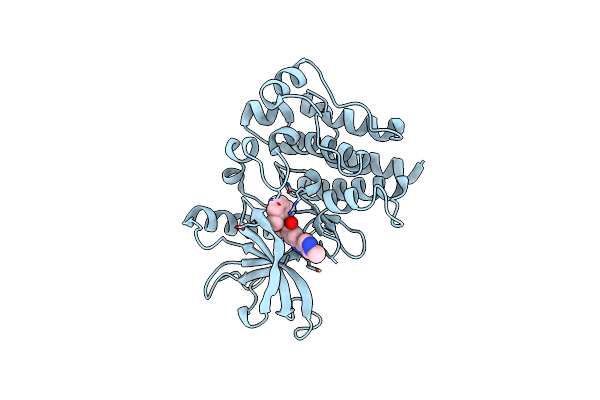 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-06-25 Classification: CELL CYCLE Ligands: A1A6M, CL, MG |
 |
Structure Of A Native Drosophila Melanogaster Pol Ii Elongation Complex Without Rpb4/Rpb7 Stalk
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Resolution:3.37 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: 9UJ, A1LYC |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.36 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: A1LYD |
 |
Cryo-Em Structure Of G1-Atpase Dimer From Mycoplasma Mobile Gliding Machinery
Organism: Mycoplasma mobile 163k
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-02-19 Classification: SIGNALING PROTEIN Ligands: GOL, A1L2E, SO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-02-19 Classification: SIGNALING PROTEIN Ligands: GOL, SO4 |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: GENE REGULATION |

