Search Count: 48
 |
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JLH, CL, IMD |
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JDF, CL, EDO, IMD, IPA, SO4 |
 |
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-10-23 Classification: VIRAL PROTEIN Ligands: ZN, SAH, EDO, IMD, YDT, CL |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Spectinomycin, Mrna, Deacylated A- And E-Site Trnaphe, And Deacylated P-Site Trnamet At 2.60A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-07 Classification: RIBOSOME/INHIBITOR,ANTIBIOTIC Ligands: MG, K, ZN, SCM, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Spectinomycin Derivative 2694, Mrna, Deacylated A- And E-Site Trnaphe, And Deacylated P-Site Trnamet At 2.75A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-08-07 Classification: RIBOSOME/INHIBITOR Ligands: MG, ZN, Y7K, SF4, K |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: A1AN8 |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: A1AON |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: 3PE, A1AOE |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: TRANSLOCASE Ligands: 3PE, A1AOF |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: MEMBRANE PROTEIN Ligands: NAG, PC1, CLR |
 |
Organism: Homo sapiens, Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: ANTIMICROBIAL PROTEIN |
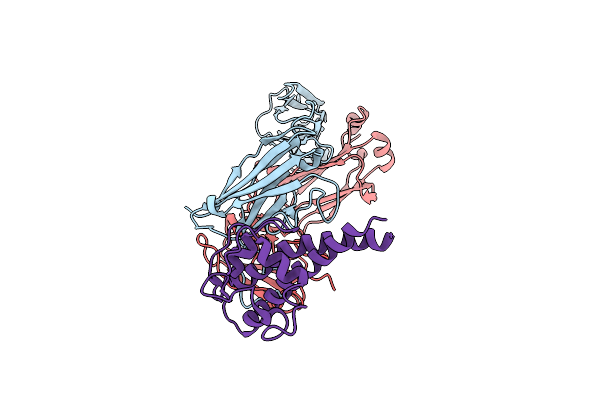 |
Organism: Pseudomonas aeruginosa, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-06-28 Classification: TRANSFERASE/TRANSFERASE Inhibitor Ligands: Y56 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2022-06-22 Classification: TRANSFERASE Ligands: AMP, P6G |
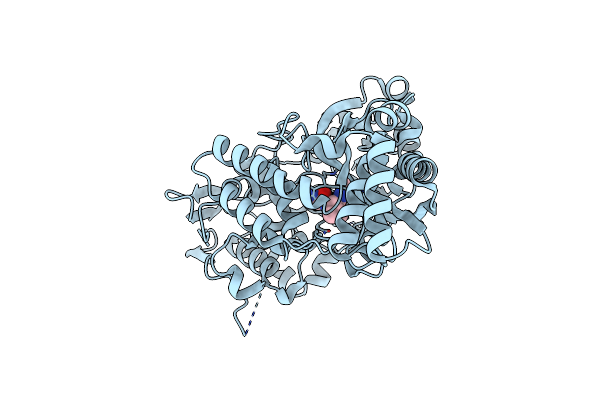 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-06-09 Classification: PEPTIDE BINDING PROTEIN |
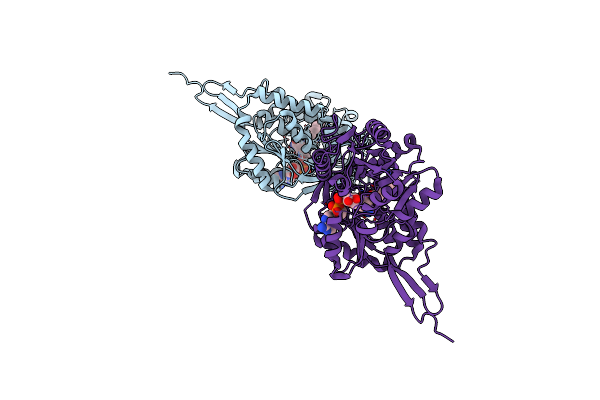 |
Crystal Structure Of Aldehyde Dehydrogenase C (Aldc) Mutant (C291A) From Pseudomonas Syringae In Complexed With Nad+ And Octanal
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: OYA, NAD |
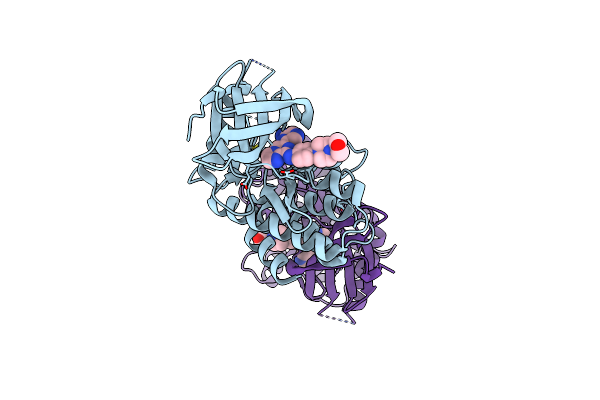 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-03-11 Classification: IMMUNE SYSTEM Ligands: R6D |
 |
Structure Of Chlamydia Trachomatis Effector Protein Cdu1 Bound To Ubiquitin
Organism: Chlamydia trachomatis 434/bu, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-08-15 Classification: HYDROLASE Ligands: CL, AYE |
 |
Structure Of Chlamydia Trachomatis Effector Protein Cdu1 Bound To Compound 5
Organism: Chlamydia trachomatis serovar l2 (strain 434/bu / atcc vr-902b)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-15 Classification: HYDROLASE Ligands: 2UQ |
 |
Structure Of Chlamydia Trachomatis Effector Protein Cdu1 Bound To Compound 3
Organism: Chlamydia trachomatis serovar l2 (strain 434/bu / atcc vr-902b)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-15 Classification: HYDROLASE Ligands: CL, D5W |

