Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
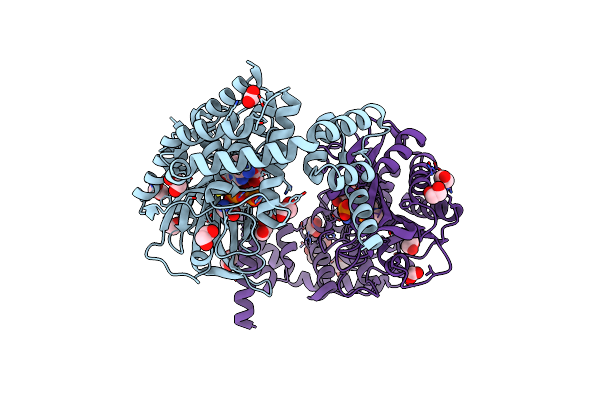 |
Crystal Structure Of Alanyl-Trna Synthetase In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, EDO, FMT, MLT, CIT |
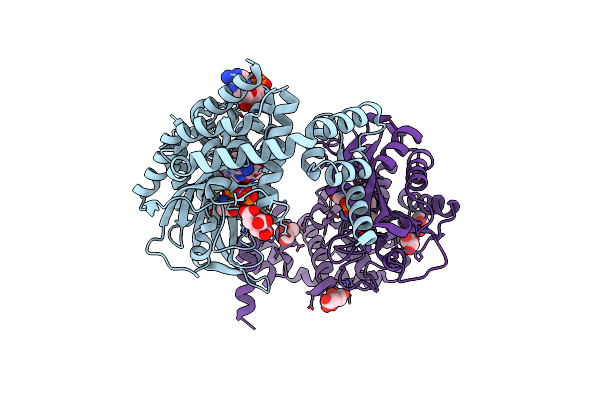 |
Crystal Structure Of Alanyl-Trna Synthetase L219M Mutant In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, CIT, TRS, GOL |
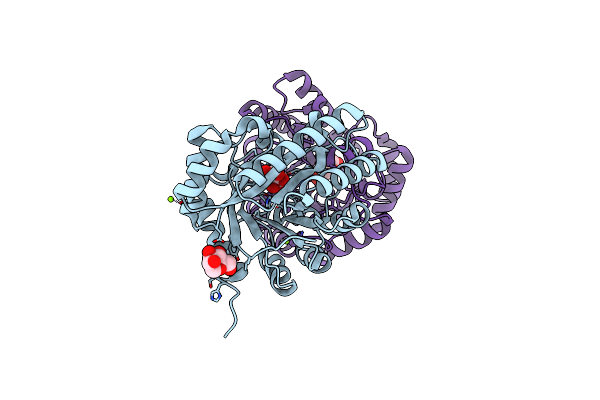 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Fructose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-28 Classification: ISOMERASE Ligands: MN, FUD, MG, BDF |
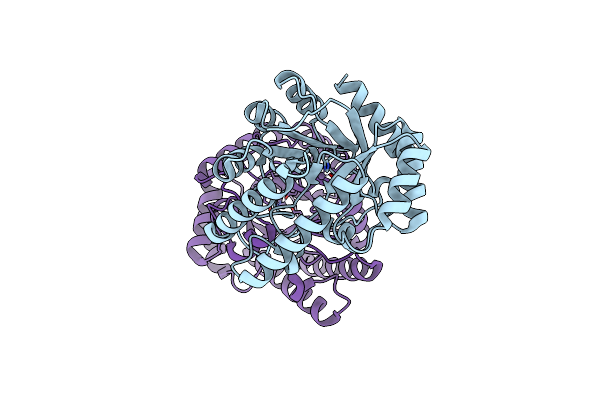 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp.
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN |
 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Allulose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, PSJ |
 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With L-Tagatose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: SOL, MN, MG, FZU, LTG |
 |
Crystal Structure Of N-Terminal His-Tagged D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Allulose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, EPE, PSJ |
 |
Crystal Structure Of N-Terminal His-Tagged D-Allulose 3-Epimerase From Methylomonas Sp. With Additional C-Terminal Residues
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, FUD, EPE, EDO |