Search Count: 6
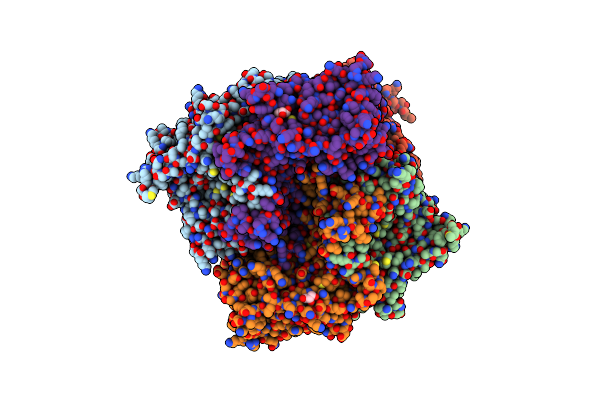 |
Crystal Structure Of Trimethylamine Methyltransferase Mttb From Methanosarcina Barkeri At 2.5 A Resolution
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: GOL, NA |
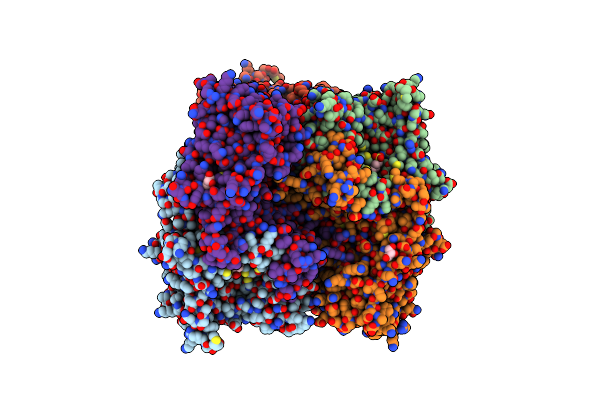 |
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: BG3, NA, GOL |
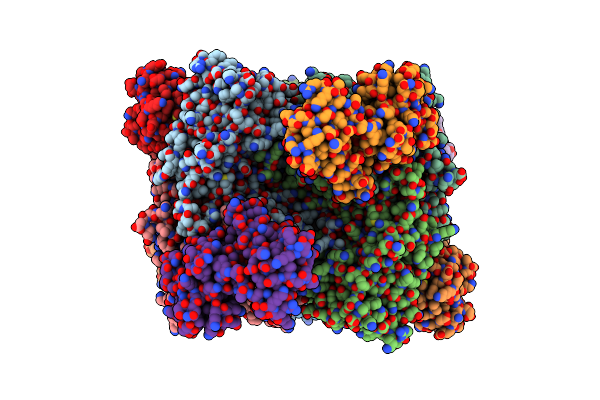 |
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: GOL, HCB |
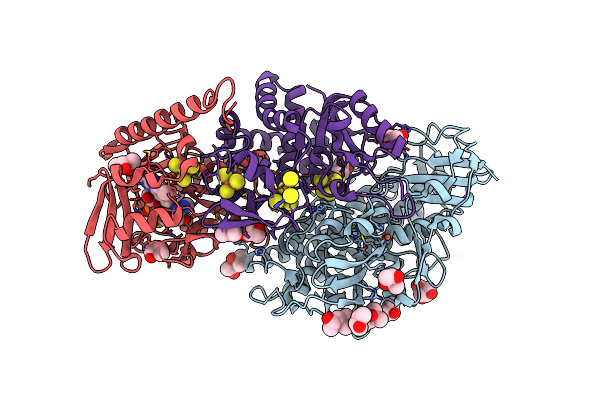 |
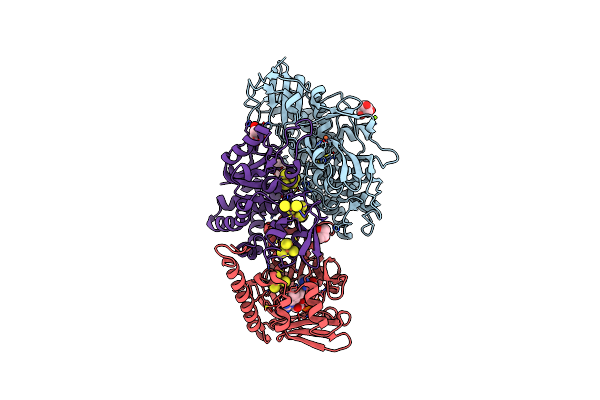
The F420-Reducing [Nife] Hydrogenase Complex From Methanosarcina Barkeri At The Nia-S State
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: SF4, FES, 144, BU3, MPD, NFU, FE, MG, FAD |
 |
The Carbon Monoxide Inhibition Of F420-Reducing [Nife] Hydrogenase Complex From Methanosarcina Barkeri
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: SF4, FES, 144, BU3, J52, FE, MG, MPD, FAD |
 |
Xenon Derivatization Of The F420-Reducing [Nife] Hydrogenase Complex From Methanosarcina Barkeri
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: SF4, 144, FES, XE, BU3, NFU, FE, MG, FAD |

