Search Count: 587
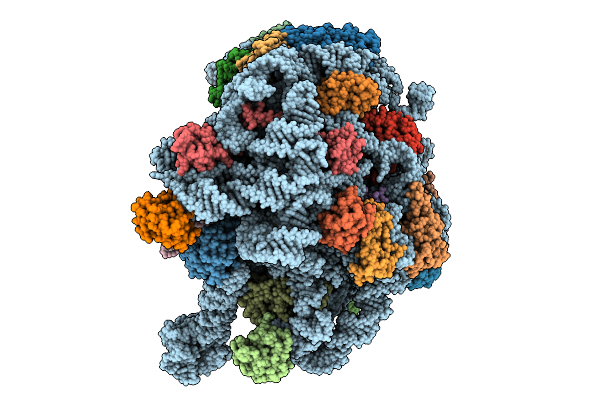 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
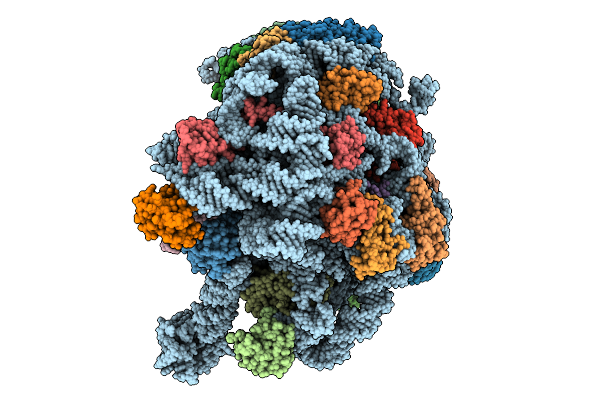 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, K, ZN |
 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
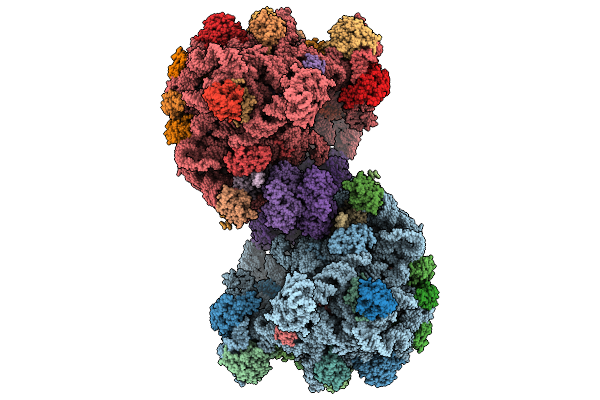 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME |
 |
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: F43, PEG, TP7, SHT |
 |
Structure Of The Native Plp Synthase Subunit Pdxs From Methanosarcina Acetivorans
Organism: Methanosarcina acetivorans c2a
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: LYASE |
 |
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-10-09 Classification: ELECTRON TRANSPORT Ligands: SF4, TPP, MG |
 |
Crystal Structure Of A Mutant Methyl Transferase From Methanosarcina Acetivorans, Northeast Structural Genomics Consortium (Nesg) Target Mvr53-11M
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: SAH |
 |
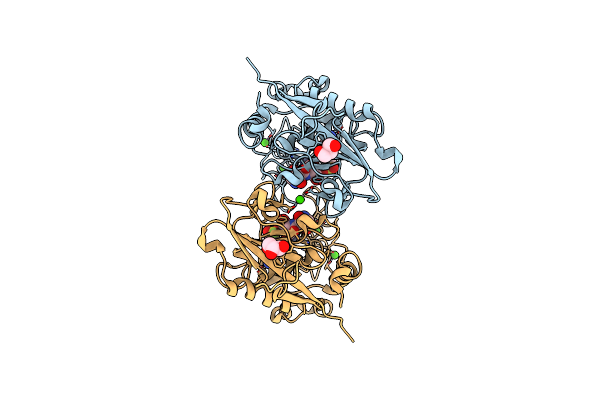
Mcrd Binds Asymmetrically To Methyl-Coenzyme M Reductase Improving Active Site Accessibility During Assembly
Organism: Methanosarcina acetivorans c2a
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: TRANSFERASE Ligands: COM, F43, TP7 |
 |
Organism: Methanosarcina acetivorans c2a
Method: ELECTRON MICROSCOPY Release Date: 2023-06-28 Classification: TRANSFERASE |
 |
Organism: Methanosarcina acetivorans c2a, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: CA, ZN, AES, GOL |
 |
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:4.50 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: ZN, CA |
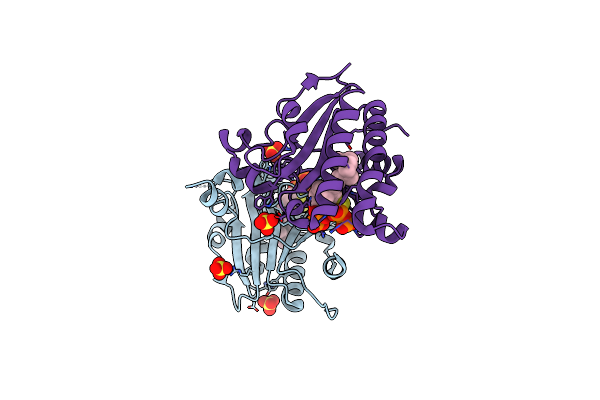 |
Structure Of Ma1831 From Methanosarcina Acetivorans In Complex With Farnesyl Thiopyrophosphate And Isopentyl S-Thiolodiphosphate
Organism: Methanosarcina acetivorans (strain atcc 35395 / dsm 2834 / jcm 12185 / c2a)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: ISY, FPS, MG, SO4 |
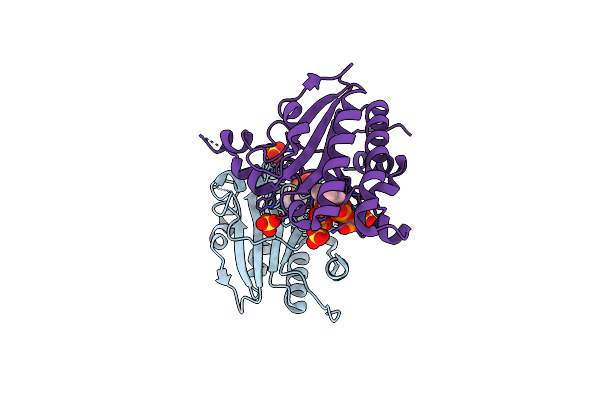 |
Structure Of Ma1831 From Methanosarcina Acetivorans In Complex With Dimethylallyl Diphosphate.
Organism: Methanosarcina acetivorans (strain atcc 35395 / dsm 2834 / jcm 12185 / c2a)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: DMA, MG, SO4 |
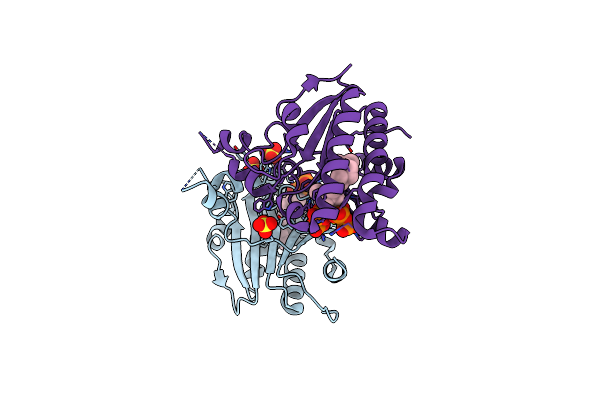 |
Structure Of Ma1831 From Methanosarcina Acetivorans In Complex With Farnesyl Pyrophosphate And Dimethylallyl Diphosphate
Organism: Methanosarcina acetivorans (strain atcc 35395 / dsm 2834 / jcm 12185 / c2a)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: DMA, FPP, MG, SO4 |
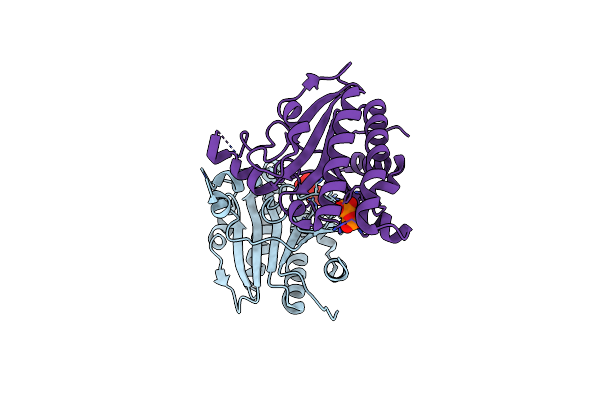 |
Structure Of Ma1831 From Methanosarcina Acetivorans In Complex With Pyrophosphate
Organism: Methanosarcina acetivorans (strain atcc 35395 / dsm 2834 / jcm 12185 / c2a)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: SO4, POP |
 |
Structure Of Ma1831 From Methanosarcina Acetivorans In Complex With Farnesyl Pyrophosphate And Geranylgeranyl Pyrophosphate.
Organism: Methanosarcina acetivorans (strain atcc 35395 / dsm 2834 / jcm 12185 / c2a)
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: FPP, SO4, BUE |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, S-Adenosyl-L-Methionine, And Peptide Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, SAM, COB, NA |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Methyl-5'-Thioadenosine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, MTA, COB, NA |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Methyl-5'-Thioadenosine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, MTA, COB, NA, PEG |

