Search Count: 17
 |
X-Ray Crystal Structure Of Methyl-Coenzyme M Reductase Glutamine Methylase (Mgma) From Methanothermobacter Marburgensis With Hydroxycobalamin
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-01 Classification: TRANSFERASE Ligands: SF4, B12, CA |
 |
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: F43, PEG, TP7, SHT |
 |
Organism: Methanosarcina acetivorans c2a
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-10-09 Classification: ELECTRON TRANSPORT Ligands: SF4, TPP, MG |
 |
X-Ray Crystal Structure Of Glycerol Dibiphytanyl Glycerol Tetraether - Macrocyclic Archaeol Synthase (Gdgt-Mas) From Methanocaldococcus Jannaschii With Archaeal Lipid, 5'Deoxyadenosine, And Methionine Bound
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-08-31 Classification: OXIDOREDUCTASE Ligands: FE, SF4, 5AD, MET, L1P, PGE, L4P |
 |
X-Ray Crystal Structure Of Glycerol Dibiphytanyl Glycerol Tetraether - Macrocyclic Archaeol Synthase (Gdgt-Mas) From Methanocaldococcus Jannaschii With Bacterial Lipid Substrate Analog, 5'Deoxyadenosine, And Methionine Bound
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-08-31 Classification: OXIDOREDUCTASE Ligands: FE, SF4, 5AD, MET, SCN, LPP |
 |
Organism: Bacillus subtilis subsp. spizizenii
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-13 Classification: LYASE Ligands: LJS |
 |
Organism: Bacillus subtilis subsp. spizizenii
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2019-02-27 Classification: LYASE Ligands: PLP, PO4, F0G |
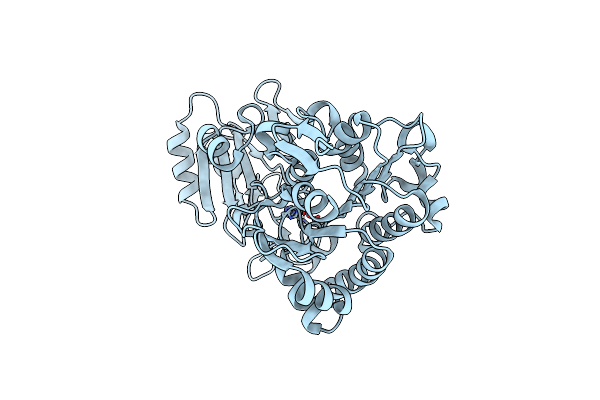 |
Crystal Structure Of Phosphonoacetate Hydrolase From Sinorhizobium Meliloti 1021 In Apo Form
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2011-08-03 Classification: HYDROLASE Ligands: ZN |
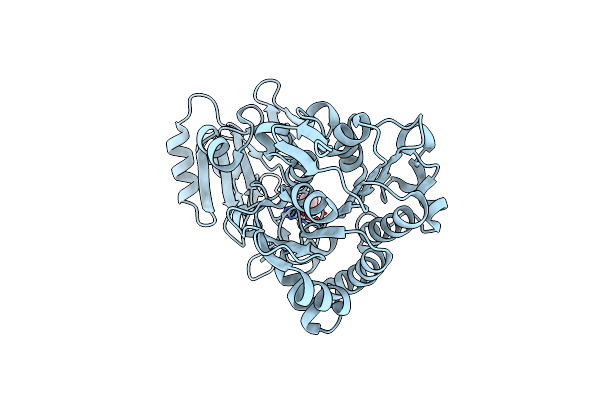 |
Crystal Structure Of Phosphonoacetate Hydrolase From Sinorhizobium Meliloti 1021 In Complex With Acetate
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-03 Classification: HYDROLASE Ligands: ZN, ACT |
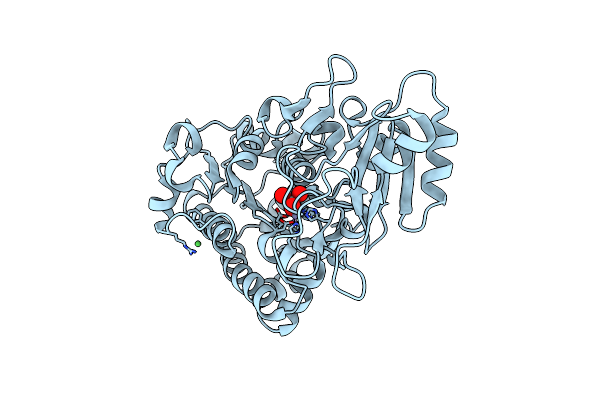 |
Crystal Structure Of Phosphonoacetate Hydrolase From Sinorhizobium Meliloti 1021 In Complex With Vanadate
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-08-03 Classification: HYDROLASE Ligands: ZN, VO4, NI |
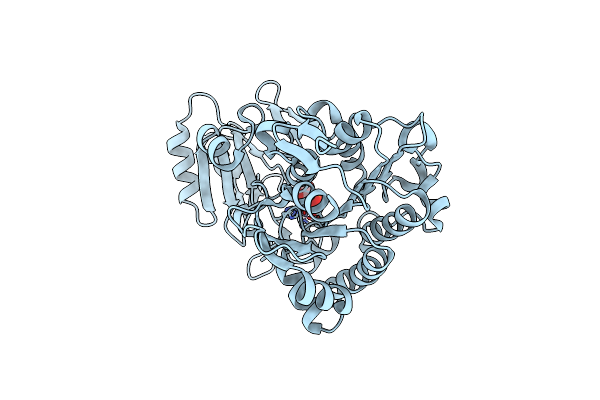 |
Crystal Structure Of Phosphonoacetate Hydrolase From Sinorhizobium Meliloti 1021 In Complex With Phosphonoformate
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, PPF |
 |
Crystal Structure Of Phosphonoacetate Hydrolase From Sinorhizobium Meliloti 1021 In Complex With Phosphonoacetate
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-03 Classification: HYDROLASE Ligands: ZN, PAE |
 |
Organism: Streptomyces luridus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-10-27 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Streptomyces luridus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-10-27 Classification: TRANSFERASE Ligands: SAM, SO4 |
 |
Organism: Streptomyces luridus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-10-27 Classification: TRANSFERASE Ligands: SAM, SO4, 2HE |
 |
Organism: Streptomyces viridochromogenes
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-06-09 Classification: BIOSYNTHETIC PROTEIN Ligands: CD |
 |
Organism: Streptomyces viridochromogenes
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2009-06-09 Classification: BIOSYNTHETIC PROTEIN Ligands: 2HE, CD |

